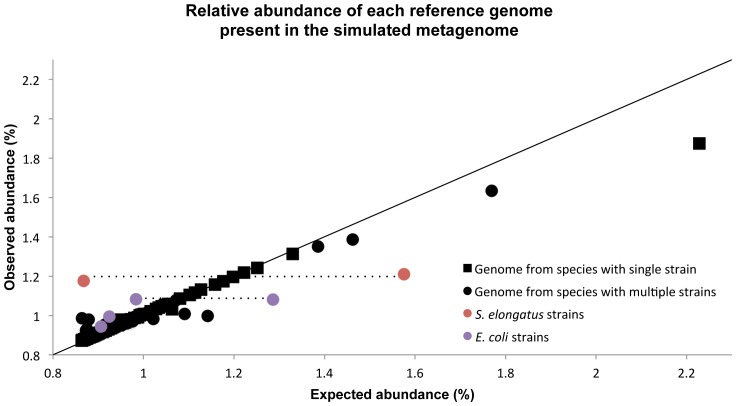
Figure 2. Relative abundance of each reference genome present in the simulated metagenome.
The observed abundances by mapping reads to reference genomes and the expected abundance correlate with a Pearson correlation coefficient of 0.95 (base and read counts). Circles represent genomes with multiple strains from one species and squares represent genomes with only one strain within the species. All, but one, of the observations deviating from the diagonal are strains from the same species. These strains are either over- or under represented because reads are mapped to other closely related strains in addition to the strain of origin. Highlighted by dashed lines, are two examples where a high sequence similarity between strains (99.9% and 98.7% for the Synechococcus elongatus and Escherichia coli strains, respectively) can result in deviations from expected abundances.