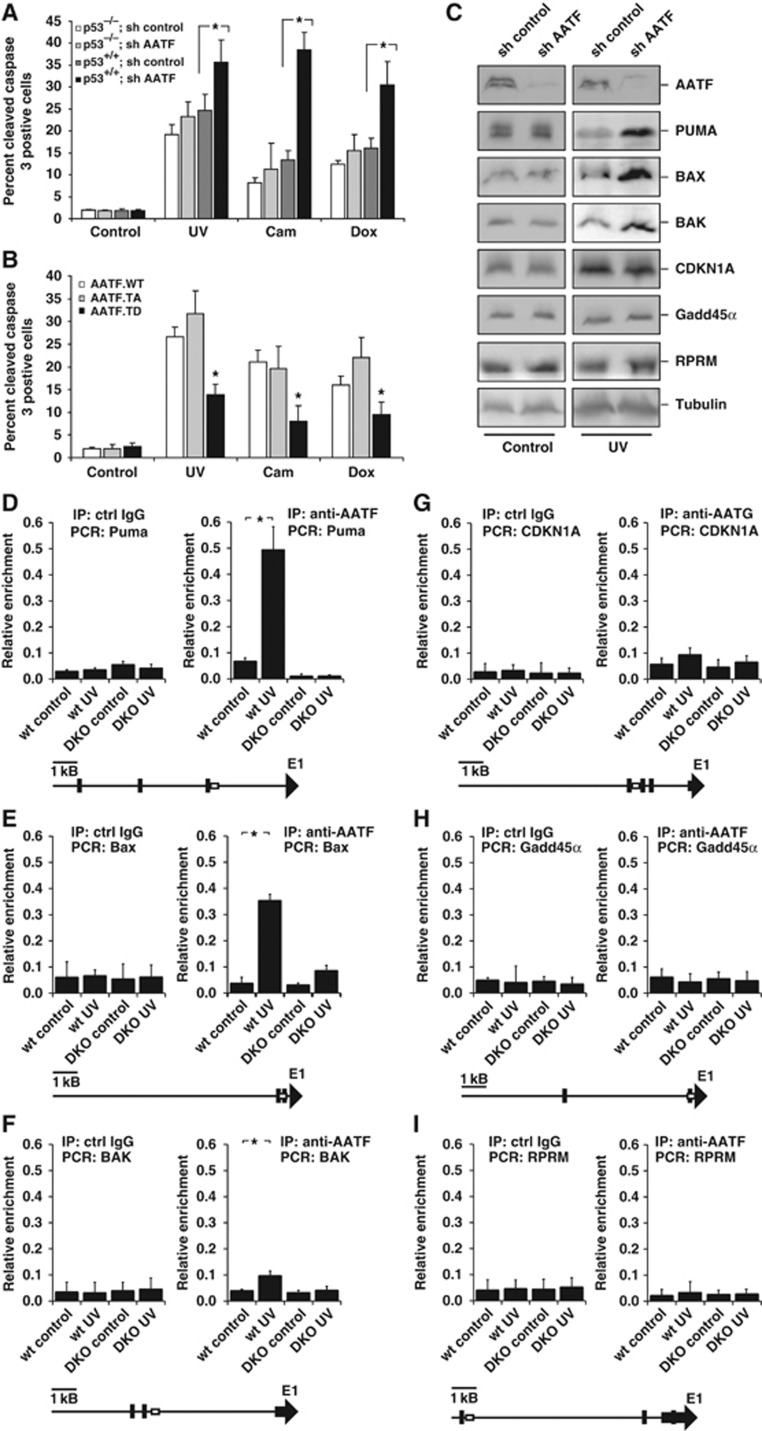
Figure 5.
AATF prevents p53-driven apoptosis by repressing DNA damage-dependent induction of pro-apoptotic genes. (A) p53+/+ and p53−/− HCT116 cells expressing control or AATF-specific shRNA were left untreated or exposed to UV (40 J/m2), camptothecin (cam, 10 μM) or doxorubicin (dox, 1 μM) and harvested for quantification of apoptosis 24 h later. AATF depletion in p53+/+ cells resulted in a significant increase in the number of apoptotic cells following all treatment regimens with DNA-damaging agents. No increase in the number of apoptotic cells could be observed in p53−/− cells following DNA damage. Asterisk indicates statistical significance, error bars represent s.d., two-tailed Student’s t-test, P<0.05, n=16. (B) p53+/+ HCT116 cells expressing AATFWT, AATFTA or AATFTD were left untreated or exposed to UV (40 J/m2), camptothecin (10 μM) or doxorubicin (1 μM) and harvested for FACS-based quantification of apoptosis 24 h later. AATFTD expression significantly repressed apoptosis in response to all three genotoxic treatments. Asterisk indicates statistical significance (P<0.05), error bars represent s.d., two-tailed Student’s t-test, P<0.05, n=16. (C) RNAi-mediated AATF depletion in p53+/+ HCT116 cells promotes DNA damage-induced induction of pro-apoptotic p53 target genes, while expression of cell-cycle-regulating p53 target genes remains unaffected. Immunoblotting was used to detect the pro-apoptotic p53-target gene products PUMA, BAX and BAK, and the cell-cycle-arresting p53 target gene products CDKN1A, GADD45α and REPRIMO 12 h after 40 J/m2 UV-C. Tubulin served as a loading control. (D–F) AATF binds to the promoters of the pro-apoptotic p53-target genes PUMA, BAX and BAK in a DNA damage and MK2-dependent manner. ChIP experiments were performed in MK2/3+/+ and MK2/3−/− MEFs that were left untreated or exposed to 40 J/m2 UV-C 60 min prior to cross-linking. DNA was precipitated using AATF antibodies. qPCR was used to quantify PUMA, BAX and BAK promoter-specific DNA in the precipitates. Unspecific IgG served as a control. Asterisk indicates statistical significance, error bars represent s.d., two-tailed Student’s t-test, P<0.05, n=9. (G–I) AATF binding to the CDKN1Ap21, GADD45α and RPRM promoter is not regulated in a DNA damage or MK2-dependent manner. ChIP experiments were performed as in (D–F) and promoter-specific primers were used for qPCR. Unspecific IgG served as a control. Asterisk indicates statistical significance, error bars represent s.d., two-tailed Student’s t-test, P<0.05, n=9. Primers to amplify the genomic DNA of the promoter regions were chosen to cover known p53-binding sites. Schematic drawings (D–I) indicated localization of primer (box) and the known p53-binding sites (black bars). Figure source data can be found with the Supplementary data.