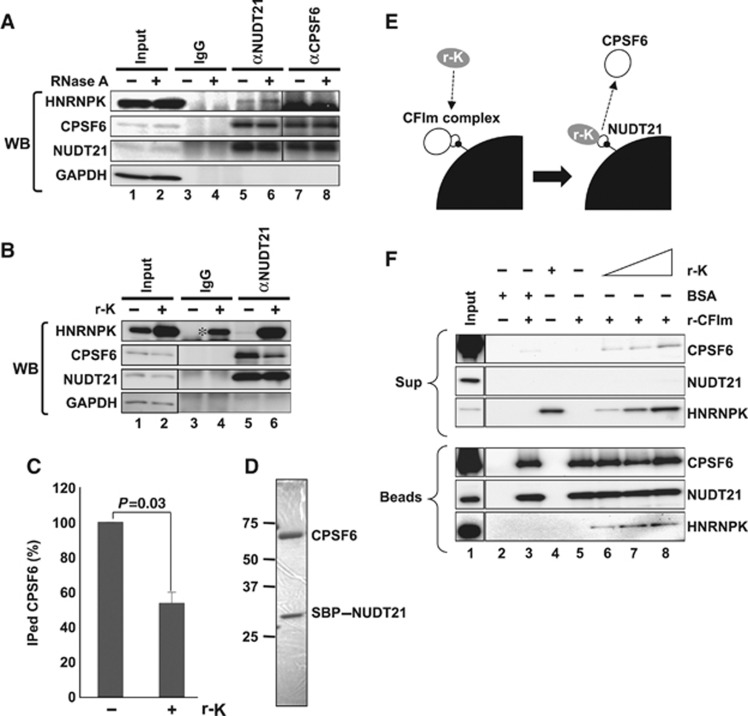
Figure 8.
HNRNPK captures NUDT21 from the functional CFIm complex. (A) Interaction between HNRNPK and NUDT21 in vivo. Immunoprecipitations with αNUDT21 and αCPSF6 were performed in the presence (+) or absence (−) of RNase A. CPSF6, NUDT21, and HNRNPK were detected by WB. (B) The CPSF6–NUDT21 interaction is affected by HNRNPK. HNE was incubated with r-K in the presence of WT RNA. Immunoprecipitation with αNUDT21 was performed in the presence (+) or absence (−) of r-K. CPSF6, NUDT21, HNRNPK, and GAPDH were detected by WB. Immunoprecipitation of CPSF6 was diminished in the presence of r-K. Asterisk represents nonspecific binding of excess r-K to IgG. (C) Quantification of immunoprecipitated CPSF6 in the presence or absence of r-K. Relative amount (%) of CPSF6 was normalized by the immunoprecipitated NUDT21 level (B). Graph shows the average (with SD) of three independent experiments. P-value was calculated by Student’s t-test. (D) Purified CFIm complex. The SDS–PAGE gel was stained with Coomassie Brilliant Blue. Both CPSF6 and SBP–NUDT21 possess Flag- and HA-tags. Size maker is shown on the left. (E) Schematics of the competitive binding assay carried out in F. The CFIm complex (30 pmol NUDT21–CPSF6 complex, white circles) was immobilized on streptavidin-conjugated beads (black quarter circle) through SBP-tag (small black circle). Recombinant HNRNPK (0–90 pmol r-K, grey circle) was mixed with the beads. Binding of r-K with NUDT21 is expected to lead to dissociation of CPSF6 from the beads. BSA (90 pmol) was used as a control. (F) Detection of CPSF6, NUDT21, and HNRNPK in bead (Beads) and supernatant (Sup) fractions. Proteins in the binding reaction are shown by+on the top of the panels. Each protein was detected by western blot, as shown on the right of the panels. Input lanes were loaded with 1/30 of the proteins used. Antibodies used are shown in Supplementary Table S4. Figure source data can be found with the Supplementary data.