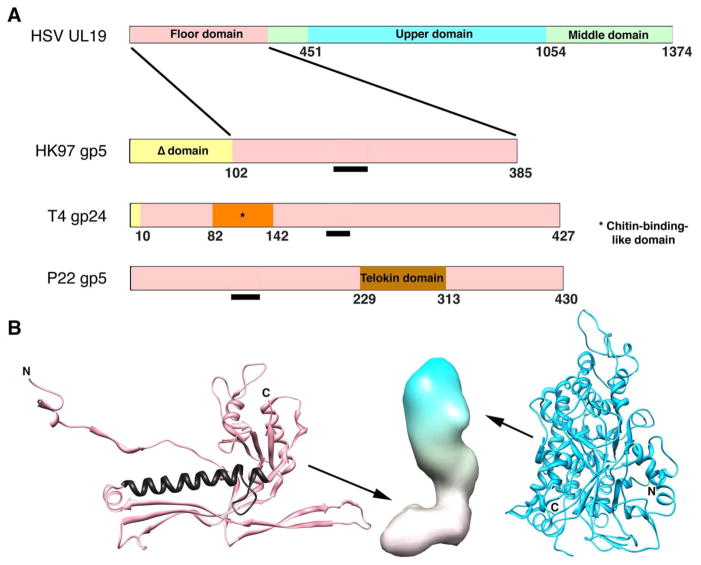
Figure 3. Domain maps of capsid proteins of HSV-1 and three bacteriophages.
(A) The phage proteins are ones for which high resolution structures have been determined. The common core domain is colored pink. The Δ-domain of HK97 is equivalent to an internal scaffolding protein fused to the core domain and is proteolyzed during maturation (gp5 -> gp5*). The T4 capsid protein gp24 forms pentamers at all non-portal vertices of this elongated T=13/Q=20 capsid, whose other capsomers are hexamers of the homolog, gp23. Its short propeptide is removed during maturation (gp24 -> gp24*). P22 does not have a protease. Gp10A of T7 (not shown) appears to have a similar core domain plus an N-terminal domain of ~ 90 residues, which is not removed by proteolysis (Agirrezabala et al. 2007). The black bars mark the location of the long α-helix, a signature feature of this fold. (B) Left: fold of HK gp5* in its mature conformation (Wikoff et al. 2000); PDB: 1OHG. The long helix is colored in black. Middle: low resolution surface rendering of a HSV-1 UL19 subunit in its B-capsid conformation. The floor domain (at bottom) is thought to have a HK97 gp5*-like fold. Right: upper domain of UL19 (Bowman et al. 2003); PDB: 1NO7.