Summary
Clathrin is a triskelion consisting of three heavy chains each with an associated light chain. During mitosis, clathrin contributes to kinetochore fibre stability. As the N-terminal domain at the foot of each leg can bind to the mitotic spindle, we proposed previously a “bridge hypothesis” wherein clathrin acts as a brace between two or three microtubules within a kinetochore fibre to increase fibre stability. Here, we have tested this hypothesis by replacing endogenous clathrin heavy chain in human cells with a panel of clathrin constructs. Mutants designed to abolish trimerization were unable to rescue the mitotic defects caused by depletion of endogenous clathrin. In contrast, stunted triskelia with contracted legs could partially rescue normal mitosis. These results indicate that the key structural features of clathrin that are necessary for its function in mitosis are a trimeric molecule with a spindle interaction domain at each end, supporting the “bridge hypothesis” for clathrin function in mitosis.
Keywords: Clathrin, mitosis, endocytosis, RNAi
Introduction
Clathrin is a three-legged molecule, or triskelion, which consists of three ~190 kDa (1,675 residue) heavy chains each with an associated ~25 kDa light chain (Kirchhausen, 2000; Kirchhausen and Harrison, 1981; Ungewickell and Branton, 1981). In mammalian cells, clathrin has two functions. First, during interphase, clathrin plays a key role in membrane trafficking (Kirchhausen, 2000). Second, when the cell enters mitosis, membrane traffic ceases (Warren, 1993) and a portion of clathrin is targeted to the mitotic spindle where it apparently stabilizes kinetochore fibres (Mack and Compton, 2001; Maro et al., 1985; Okamoto et al., 2000; Royle et al., 2005; Sutherland et al., 2001). When clathrin heavy chain (CHC) is depleted from cells using RNAi, a number of mitotic defects arise such as problems in congression (the movement of chromosomes to the metaphase plate), destabilization of kinetochore fibres and lengthened mitosis due to prolonged signalling of the spindle checkpoint (Royle et al., 2005).
The organization of a clathrin triskelion, as defined by a recent molecular model (Fotin et al., 2004) is shown in Fig. 1A. A single CHC molecule consists of an N-terminal seven-bladed β-propeller, a linker region, eight clathrin heavy chain repeat (CHCR0-7) segments, a proximal hairpin, a tripod region that is thought to be responsible for trimerization, and a variable C-terminal segment (residues 1631-1675). Thus, one CHC molecule resembles a human leg: the foot comprises the N-terminal domain, linker and part of CHCR0; the ankle corresponds to the remainder of CHCR0, CHCR1 and CHCR2; and the knee is at CHCR5 (Fotin et al., 2004).
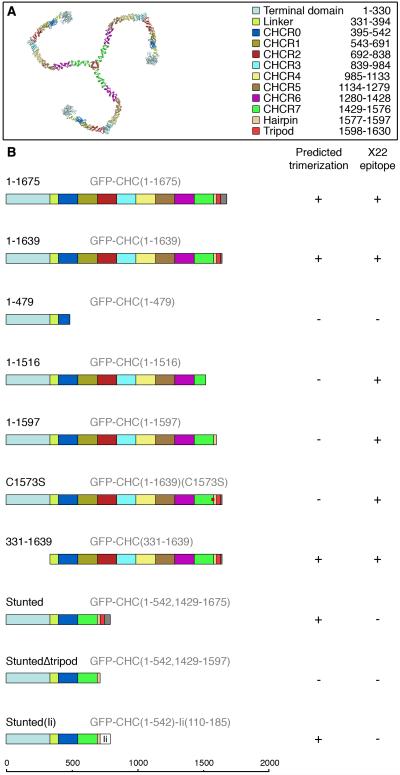
Figure 1. Overview of the organisation of clathrin and of the constructs used in this study.
(A) Model of a clathrin triskelion proposed by Fotin et al. 2004. The triskelion is viewed looking down onto the vertex. Coloured regions show the features of a CHC molecule (see key, right).
(B) Schematic representations of each CHC construct used in the study. Variable region (residues 1631-1675) is shown in grey, GFP has been omitted for clarity. Short names used in the paper are in black and full descriptive names are in grey. Trimerization was predicted based on previous publications (Fotin et al., 2004; Liu et al., 1995; Nathke et al., 1992; Ybe et al., 2003). X22 epitope is between residues 1109-1128 of CHC (Liu et al., 1995). Constructs were compared to ‘GFP’, GFP expressed on a CHC RNAi background and to ‘Control’, GFP expressed on a control RNAi background.
As the N-terminal domain at the end of each leg can bind to the mitotic spindle, we proposed a “bridge hypothesis” wherein clathrin triskelia act as a brace between two or three microtubules within a kinetochore fibre to increase fibre stability (Royle et al., 2005). An alternative view is that clathrin does not act as a bridge, but as a lattice or matrix that can support spindle fibres. In our earlier paper (Royle et al., 2005), we showed that normal mitosis could be rescued by full-length clathrin triskelia and not by the N-terminal domain alone, but this did not allow us to distinguish between these two models.
In the present study, we set out to test these two hypotheses by replacing endogenous clathrin heavy chain (CHC) in human cells with a variety of CHC constructs. These constructs allowed us to ask: is trimerization essential for the function of clathrin in mitosis? And what are the minimal structural requirements for normal mitosis? Our findings exclude the “lattice” model and support the “bridge hypothesis” for clathrin function in mitosis.
Results
To test whether or not the triskelion structure of clathrin was essential for its function in mitosis, we designed a panel of clathrin constructs based on structural (Fotin et al., 2004) and biochemical information (Liu et al., 1995; Nathke et al., 1992; Ybe et al., 2003). These various constructs were expressed in HEK293 cells in which levels of endogenous CHC were reduced by more than 90% using RNA interference (RNAi).
An introduction to the constructs used in this study
The CHC constructs used in this study are illustrated in Fig. 1B. The first two constructs have the trimerization domain and should be able to form trimers: full-length CHC (1-1675) and the major splice variant (1-1639). Four other constructs are all predicted to be unable to trimerize: three truncations (1-479, 1-1516, 1-1597) and a point mutant (C1573S). We also included a construct that is predicted to trimerize but lacks the N-terminal domain (331-1639) in order to test the role of the β-propeller interaction domain. Note that our earlier analysis was limited to 1-1639 and 1-479 only (Royle et al., 2005).
All CHC constructs were GFP-tagged at the N-terminus and any that included CHC residues 60-66 (the region targeted for RNAi) were rendered resistant to knockdown (see Materials and Methods). For comparison we expressed GFP alone on a CHC-depleted background (GFP) or as a control we expressed GFP alone on an endogenous clathrin background (Control).
As the constructs were expressed on a CHC RNAi background, we first assessed the level of CHC in these cells by immunocytochemistry using the monoclonal antibody, X22 (Fig. 2). We found that in GFP cells the level of endogenous clathrin was ~10% of that in Control (Royle et al., 2005). Knockdown occurred to a similar extent in cells expressing 1-479. In cells expressing 1-1675, 1-1639, 1-1516, 1-1597, C1573S and 331-1639, X22 recognised the expressed protein (Fig. 2, Supplementary Fig. 1). This is consistent with previous studies that mapped the epitope for this antibody to residues 1109-1128 of CHC (Liu et al., 1995). We can be confident that knockdown actually occurred in these cells because we saw differential effects on clathrin-mediated endocytosis (CME) and mitotic rescue.
Figure 2. Knockdown of endogeneous CHC and replacement with GFP-tagged CHC constructs.
(A) Representative confocal images of cells expressing CHC constructs (left, green) that were immunostained for CHC (middle, red) using antibody X22. Nucleic acids are shown in blue in the merged panels (right). Scale bar, 10 μm. Note the knockdown of endogenous CHC in GFP and 1-479 and the X22 immunoreactivity in 1-1675.
(B) Histogram to compare the amount of X22 immunoreactivity in cells expressing each of the CHC constructs. Cells were outlined using the GFP channel as a guide and the mean greyscale pixel value for the red signal (X22/Alexa546) was measured. Results are mean ± s.e.m., from 26-47 cells per construct and are normalized to Control.
Stunted constructs were designed to mimic the structural features of CHC
We also wanted to ask: what are the minimal structural requirements for the function of clathrin in mitosis? To address this point we designed a construct in which the foot of CHC was grafted onto the thigh (Stunted) to test whether this protein could recapitulate the proposed structural role of clathrin in mitosis according to the “bridge hypothesis”, i.e. a trimeric molecule with an interaction domain at each end (Fig. 1B). We designed two further constructs: a trimerization-deficient version of Stunted that lacks the trimerization domain (StuntedΔtripod) and a variation of Stunted, designated Stunted(Ii), that instead uses the C-terminal trimerization domain from the invariant chain of MHC II (Wakeham et al., 2003). Also, the three Stunted constructs lack CHCR1-6, which are necessary for lattice formation (Fotin et al., 2004). When cells expressing either Stunted, StuntedΔtripod or Stunted(Ii) were stained for X22, we found that endogenous CHC was depleted to <10% of Control and there was no recognition of the expressed protein (Fig. 2 and Supplementary Fig. 1).
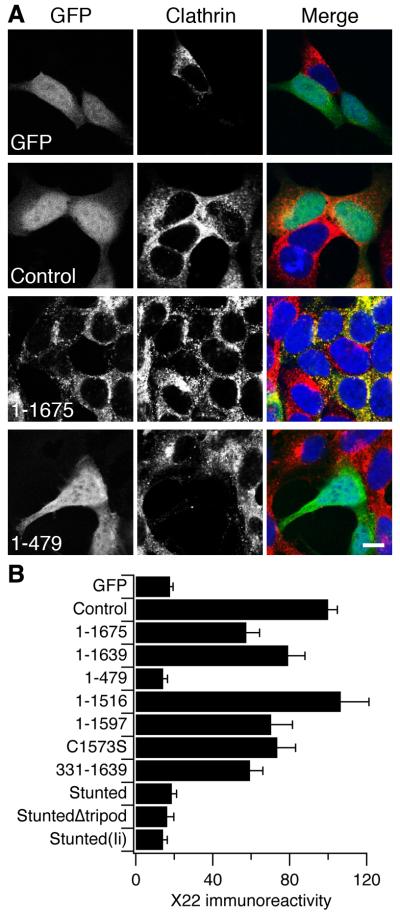
Although Stunted was engineered to form small triskelia, it was important to verify that this construct could actually form trimers. Previously, analytical ultracentrifugation has been used on bacterially-expressed CHC constructs to check their oligomeric state (Wakeham et al., 2003). In that study, trimeric CHC constructs gave a “dual peak” profile that corresponded to monomers and trimers. Here we used a simple hydrodynamic method to assess the oligomeric state of our CHC constructs expressed in HEK293 cells that were depleted of endogenous CHC. Fig. 3A shows the results from a typical sedimentation analysis experiment where 1-1639 and 1-1597 were separated on a 15-40% glycerol gradient. For 1-1639, two clear peaks could be distinguished which corresponded to weight expected for monomers and trimers. The second peak was effectively eliminated when the tripod domain is removed (1-1597). The “dual peak” profile for 1-1639 was similar to the results of Wakeham et al. (2003) and therefore gave us a fingerprint for recognising a trimeric molecule. When Stunted was separated on a 10-35% glycerol gradient, two clear peaks could also be distinguished in the fractions corresponding to the mass expected for monomers and trimers (Fig. 3B). When StuntedΔtripod was run on a 10-35% gradient, the second peak was eliminated. These observations suggest that Stunted does indeed exist as a trimeric molecule.
Figure 3. Stunted is a trimeric molecule.
(A) Sedimentation analysis of 1-1639 and 1-1597. Quantification of a typical experiment where cell lysates were separated on a 15-40% glycerol gradient. Note the second peak at fraction 8 in 1-1639 that is absent in 1-1597.
(B) Sedimentation analysis of Stunted and StuntedΔtripod. Quantification of a typical experiment where cell lysates were separated on a 10-35% glycerol gradient. Note the presence of a second peak in Stunted that is absent in StuntedΔtripod. The sedimentation of molecular weight standards is shown for each gradient (arrows). The calculated molecular weights for monomers of 1-1639, 1-1597, Stunted and Stunted Δtripod were 215, 210, 117 and 108 kDa respectively. Fractions were analysed by SDS-PAGE and immunoblotting for GFP. Following densitometry, the signal in each fraction was expressed as a percentage of the total GFP signal from all fractions on the blot.
Rescue of clathrin-mediated endocytosis by CHC constructs
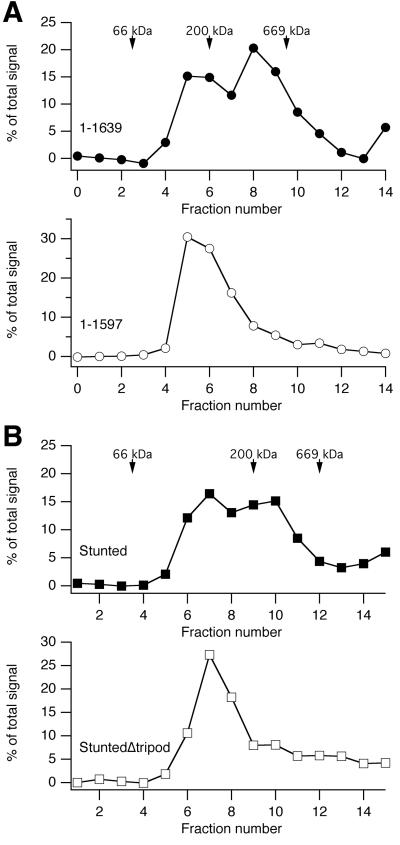
Before testing for rescue of mitosis, we first broadly characterised our panel of CHC constructs. To test the function of CHC constructs in CME, we assayed uptake of transferrin-Alexa546 by confocal microscopy (Fig. 4). Transferrin uptake was 10.2 ± 1.9% in GFP cells compared to Control. With this result in hand we could then test which constructs could rescue normal CME. Of the CHC constructs, only 1-1675 and 1-1639 supported normal transferrin uptake (Fig. 4). None of the remaining constructs supported CME. CHC constructs 1-479, 1-1516, 1-1597, C1573S and StuntedΔtripod are all predicted to be impaired in trimerization and so would be unable to form clathrin triskelia. 331-1639, Stunted and Stunted(Ii) are predicted to be trimeric, but would not be able to form functional triskelia; because Stunted and Stunted(Ii) lack CHCR1-6 which are essential for polymerisation and because 331-1639 lacks the N-terminal region that is needed to interact with AP2 (Murphy and Keen, 1992; Shih et al., 1995) in order for CME to occur.
Figure 4. CME was only supported in cells expressing functional triskelia.
(A) Representative confocal micrographs of transferrin uptake in cells expressing some of the CHC constructs on a CHC knockdown background. Panels show GFP (green, left), transferrin-Alexa546 (red, middle) and merge (right). Scale bar, 20 μm.
(B) Histogram to show quantification of transferrin uptake. Each cell was outlined using the GFP channel as a guide, the transferrin-Alexa546 image was then thresholded and the number of puncta within the ROI was counted. Results are mean ± s.e.m., from 18-38 cells per construct, **, p < 0.01, normalized to Control.
Spindle recruitment of CHC constructs
As part of our characterisation of the CHC constructs, we next assessed the subcellular distribution of each construct. In interphase cells, 1-1675, 1-1639, C1573S and 331-1639 were distributed in numerous puncta similar to GFP-LCa or endogenous clathrin; 1-479 had a diffuse cytosolic distribution with no puncta whereas the remainder had a punctate accumulation in a perinuclear compartment but no other puncta within the cell (data not shown).
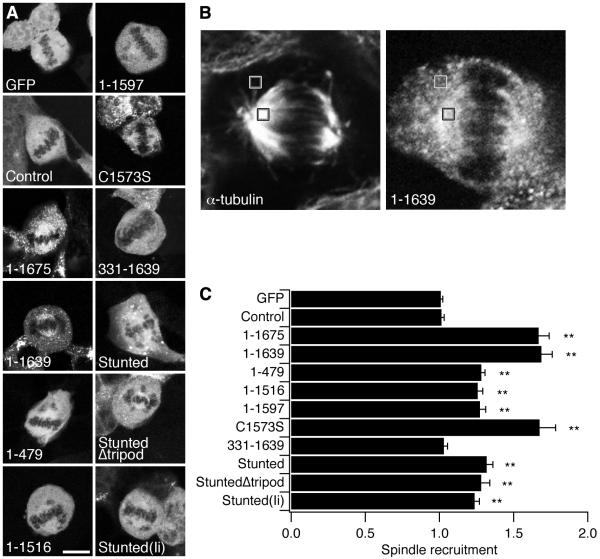
In order for a given construct to act as a bridge or a lattice at the mitotic spindle, it must be targeted to the spindle to some degree. We therefore measured the recruitment of each CHC construct to the mitotic spindle using a fluorescence-based method (Royle et al., 2005). In this assay, the GFP fluorescence of a given protein on the spindle is compared to that in the cytoplasm (Figs 5A and B). Both GFP and Control had a spindle recruitment ratio of approximately 1 indicating no recruitment (Fig. 5C). We found weak recruitment (between 1.24 and 1.32) for constructs that included the N-terminal β-propeller: 1-479, 1-1516, 1-1597, Stunted, Stunted(Ii) and StuntedΔtripod. We measured no enrichment of 331-1639 at the spindle (1.03 ± 0.03), again suggesting that the β-propeller domain at the foot of the triskelion is necessary for binding the spindle. We saw stronger recruitment for 1-1675, 1-1639 and C1573S (around 1.67). In summary, all CHC constructs except 331-1639 were recruited to the mitotic spindle.
Figure 5. Constructs containing the N-terminal domain were recruited to the mitotic spindle.
(A) Representative confocal micrographs of each construct in HEK293 cells in metaphase. Scale bar, 10 μm.
(B) A cell expressing 1-1639 on a knockdown background. ROIs (grey, 1.04 × 1.04 μm) were placed over the spindle (black outline) and in the cytoplasm (white outline). Spindle regions and non-spindle areas were defined by staining for α-tubulin using DM1A/Alexa546 (left). Measurements were taken from the GFP channel (right).
(C) Histogram to show the recruitment to the spindle of CHC constructs on a CHC knockdown background. Spindle recruitment is the GFP fluorescence measured in a 1.04 × 1.04 μm ROI at the spindle divided by that measured in a same-sized ROI in the cytoplasm (Fspindle/Fcytoplasm). Results are mean ± s.e.m., from 12-27 cells per construct, **, p < 0.01.
Rescue of mitotic defects by CHC constructs
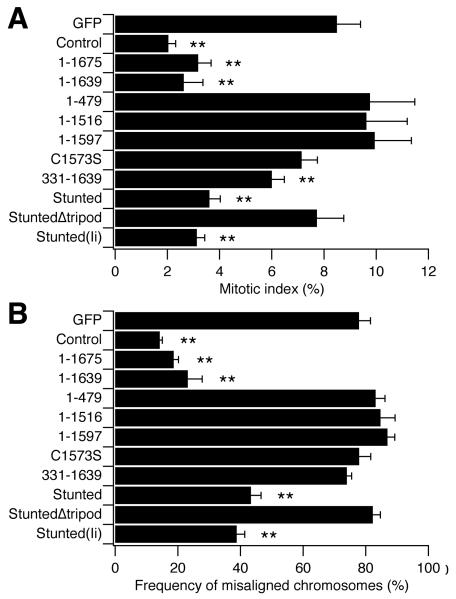
When CHC is depleted from cells using RNAi, a number of mitotic defects arise such as defective congression, destabilization of kinetochore fibres and lengthened mitosis due to prolonged signalling of the spindle checkpoint (Royle et al., 2005). We next tested the ability of these proteins to rescue the mitotic defects that result from depletion of endogenous CHC. We first examined the mitotic index, a measure of the proportion of cells undergoing mitosis, and therefore an indicator of time spent in mitosis (Fig. 6A). In Control cells the mitotic index was 2 ± 0.3% and in GFP cells, where CHC was depleted, mitosis was four-fold longer (8.5 ± 0.9%). Mitotic index was rescued in cells expressing 1-1675 and 1-1639, and was rescued partially in cells expressing Stunted, Stunted(Ii) and 331-1639. In contrast, the constructs with disrupted trimerization (1-479, 1-1516, 1-1597, C1573S and StuntedΔtripod) all failed to reduce the time spent in mitosis (Fig. 6A). These results are in agreement with the idea that trimerization is essential for the function of clathrin in mitosis. Furthermore, the partial rescue of mitosis by Stunted, Stunted(Ii), but not StuntedΔtripod, argued for a bridging function for clathrin. However the slight reduction of mitotic index by 331-1639 (from 8.5% to 5.6 ± 0.6%) was unexpected, because this protein lacks the N-terminal β-propeller domain that is necessary for binding to the mitotic spindle; it was not recruited to the spindle (Fig. 5C) and should therefore be unable to bridge microtubules.
Figure 6. Rescue of mitotic defects by trimeric CHC constructs.
Histograms to show quantification of (A) mitotic index and (B) the frequency of metaphase-like cells with misaligned chromosomes in cells expressing CHC constructs on a CHC knockdown background. Mitotic index was assessed by counting the number of GFP-positive cells in mitosis as a proportion of the total number of GFP-positive cells, per unit area. The frequency of GFP-positive cells in a metaphase-like state with misaligned chromosomes were also counted. H33342 staining was used to identify mitotic cells and misaligned chromosomes. Results are mean ± s.e.m., **, p < 0.01.
In order to verify the experiments that measured mitotic index, we examined a second quantifiable defect in mitosis: the frequency of metaphase-like cells with misaligned chromosomes (Fig. 6B). In Control cells the frequency was 14.2 ± 0.8% compared to 77.8 ± 3.6% in GFP cells. Normal alignment of chromosomes was rescued in cells expressing 1-1675 and 1-1639 and was rescued partially in cells expressing Stunted and Stunted(Ii) only. The trimerization mutants (1-479, 1-1516, 1-1597, C1573S and StuntedΔtripod) were again ineffective at rescuing this mitotic defect. In contrast to the mitotic index measurements, cells expressing 331-1639 had a frequency of metaphase-like cells with misaligned chromosomes of 75.2 ± 2%, indicating no rescue. We also examined kinetochore-spindle contacts in metaphase-like cells following depolymerisation of non-stable kinetochore fibres (Supplementary Fig. 2, Yao et al., 2000). This is a qualitative assay of kinetochore fibre stability that has been used previously (Royle et al., 2005). We found that for Control, 1-1675, 1-1639, Stunted and Stunted(Ii), all kinetochores had stable fibre attachments; whereas for GFP, 1-479, 1-1516, 1-1597, C1573S, 331-1639 and StuntedΔtripod, orphan kinetochores were frequently found. These results were in keeping with those in Fig. 6. We conclude that 331-1639 is not competent to function in mitosis because although there was a slight reduction of mitotic index, we saw no rescue of either the frequency of misaligned chromosomes or the stability of kinetochore fibres in cells expressing 331-1639.
We finally wanted to check whether or not the differences in functional rescue could be attributed to differences in expression. For example, the lack of rescue with 1-1597 may be due not to its lack of trimerization, but to a lower expression level than 1-1639. The levels of expression for each construct were assessed by measuring GFP fluorescence (Supplementary Fig. 3). Expression was variable, ranging from ~40% to ~90% of Control. There was no correlation between expression levels and functional rescue. The expression of constructs that rescued mitosis ranged from 40% to 72%, whereas those without effect varied from 38% to 80%. These results ruled out poor expression as an explanation for failure of 1-479, 1-1516, 1-1597, C1573S, 331-1639 and StuntedΔtripod to rescue mitosis; leading us to conclude that it was their lack of trimerization that was responsible for a lack of functional rescue.
Discussion
The apparent function of clathrin in mitosis is to stabilize the fibres of the mitotic spindle. We had shown previously that depletion of endogenous CHC resulted in defects in mitosis and that clathrin triskelia but not the isolated N-terminal domain could rescue these defects (Royle et al., 2005). Two alternative hypotheses arose out of these observations. The “bridge hypothesis” for the function of clathrin in mitosis suggests that CHC connects microtubules because the feet of a triskelion act as attachment points and trimerization at the vertex forms a rigid connection. Whereas the “lattice hypothesis” suggests that clathrin triskelia form a lattice-like matrix which can support the fibres of the mitotic spindle (Scholey et al., 2001). In the present study we were able to distinguish between these two models.
Our results (summarized in Supplementary Table 1) show that trimerization of clathrin is essential for the normal function of clathrin in mitosis because trimerization-deficient CHC constructs were unable to rescue normal mitosis, whereas constructs that were able trimerize could support mitosis. In addition, we found a partial rescue of normal mitosis by Stunted and Stunted(Ii), but not StuntedΔtripod. These observations illustrate that the minimal structural requirements for the function of clathrin in mitosis are a trimeric molecule that has a β-propeller domain at each end. As the Stunted constructs lack CHCR1-6, which are necessary for lattice formation, then the partial rescue of mitosis with these constructs must be due to the molecules acting as bridges and not as a lattice. Together our results exclude the “lattice hypothesis” and provide strong support for the “bridge hypothesis” of the function of clathrin in mitosis, wherein clathrin acts as a three-legged brace between two or three microtubules within a spindle fibre to increase fibre stability.
A single fibre of the mitotic spindle comprises many microtubules and in order to give strength and stability to the fibre as it manoeuvres chromosomes around the cell, the microtubules are cross-linked by electron-dense material (Compton, 2000). Besides clathrin, other cross-linking molecules have been described. For example, a bipolar molecule, motor KLP61F, has been proposed to cross-link microtubules in interpolar microtubule bundles (Sharp et al., 1999) and a recent report has suggested that NuSAP may also bridge microtubules, although its multimerization state is unknown (Ribbeck et al., 2006). Is the trimeric structure of clathrin best suited to bridging spindle fibres? The partial rescue of mitosis with Stunted and Stuned(Ii), but not StuntedΔtripod was intriguing because it suggested that although the β-propeller domains must be trimerized in order for kinetochore fibres to be stabilized, it was less important how they were trimerized. Stunted had the trimerization domain from CHC whereas Stunted(Ii) had the C-terminal trimerization domain of an unrelated protein (CD74 antigen invariant chain residues 110-195). Whether or not dimerized or tetramerized CHC feet can also rescue mitosis is an interesting question for the future. Perhaps a dimer actually constitutes a better design for a bridge, but reusing a three-legged molecule that is suited to endocytosis was the best solution to stabilizing spindle fibres that evolution could provide.
CHC constructs that contained the N-terminal domain were enriched at the mitotic spindle, in keeping with the idea that the feet of clathrin triskelia constitute the attachment points for clathrin at the mitotic spindle. However we measured more prominent spindle recruitment for 1-1675 and 1-1639, compared to 1-1597, suggesting that other C-terminal sequences may somehow regulate binding to the mitotic spindle. In addition, C1573S had a similar subcellular distribution as 1-1675 and was recruited to the spindle to a similar degree. This was unexpected because as a trimerization-deficient construct (Ybe et al., 2003), C1573S would be predicted to behave similar way to 1-1597. Clearly, it is not merely trimerization that causes increased recruitment because C1573S was recruited similarly to 1-1675, while Stunted and Stunted(Ii) were not. Perhaps C-terminal residues between 1597 and 1639 are important for spindle binding, but only in the context of a full-length clathrin leg.
While our results argue for the “bridge hypothesis” and against the “lattice hypothesis”, another possibility remains. Clathrin may recruit to the spindle another protein that can mediate fibre stability. In this scheme, trimeric clathrin constructs that contain the β-propeller domain bind the partner with more avidity and can therefore rescue mitosis in the absence of endogenous clathrin. It is therefore important to not only identify the protein partners that recruit clathrin to the mitotic spindle but to determine whether or not clathrin recruits any proteins to the spindle with the ability to stabilise spindle fibres.
An important next step in further elucidating the function of clathrin in mitosis will be to better understand the interactions between the feet of the triskelion and the spindle. How is the bridging function regulated? What protein(s) are involved in targeting clathrin to the mitotic spindle and in segregating clathrin from microtubules during interphase?
Materials and Methods
Molecular biology
DNA plasmids to simultaneously knockdown endogenous human CHC by RNAi through expression of shRNA and to express fluorescent proteins under a CMV promoter, dubbed pBrain constructs, were described previously (Royle et al., 2005). Human CHC was knocked down using CHC4 shRNA. A control shRNA (CHC1) targeted rat CHC and was ineffective in knocking-down expression of human CHC. The plasmids pBrain-GFP-CHC1 (Control), pBrain-GFP-CHC4 (GFP), pBrain-GFP-CHC(1-479)KDP-CHC4 (1-479) and pBrain-GFP-CHC(1-1639)KDP-CHC4 (1-1639) were all available from previous work (Royle et al., 2005). To make 1-1675, an Asp718-Sac II fragment from the full-length human CHC cDNA (Kazusa KIA 00034) was inserted into a vector containing GFP-CHC(1639)KDP and then an ApaL I-ApaL I fragment from pBrain-GFP-CHC(1-1639)KDP-CHC4 was inserted to give pBrain-GFP-CHC(1-1675)KDP-CHC4.
The truncations 1-1516 and 1-1597, were generated by first inserting PCR fragments with premature stop codons into Xba I-Sac II sites of GFP-CHC(1675)KDP and then an ApaL I-ApaL I fragment from pBrain-GFP-CHC(1-1639)KDP-CHC4 was inserted to give pBrain-GFP-CHC(1-1516)KDP-CHC4 and pBrain-GFP-CHC(1-1597)KDP-CHC4.
Stunted was made by first inserting an Asp718-Sac II PCR fragment into GFP-CHC(1-479)KDP (where the Sac II site encodes residues 542 and 1429) to give GFP-CHC(1-542)KDP, then inserting a Sac II-BamH I PCR fragment to make GFP-CHC(1-542,1429-1675)KDP, finally an ApaL I-Bgl II fragment from pBrain-GFP-CHC4 was inserted to give pBrain-GFP-CHC(1-542,1429-1675)KDP-CHC4.
StuntedΔtripod was made by inserting a Sac II-Sac II PCR fragment into GFP-CHC(1-542)KDP and to make GFP-CHC(1-542,1429-1597)KDP, and an ApaL I-Bgl II fragment from pBrain-GFP-CHC4 was inserted to give pBrain-GFP-CHC(1-542,1429-1597)KDP-CHC4.
To make Stunted(Ii), the C-terminal trimerization domain of CD74 antigen (invariant polypeptide of MHC II antigen associated) I.M.A.G.E. 4853578 was amplified by PCR and inserted at Sac II-BamH I of GFP-CHC(1-542)KDP to make GFP-CHC(1-542)KDP-Ii(110-185), and an ApaL I-Bgl II fragment from pBrain-GFP-CHC4 was inserted to give pBrain-GFP-CHC(1-542)KDP-Ii(110-185)-CHC4.
The terminal domain truncation construct 331-1639 was made by inserting a Bgl II-Afl III fragment from GFP-CHC(331-1074) into pBrain-GFP-CHC(1-1639)KDP-CHC4 to give pBrain-GFP-CHC(331-1639)-CHC4.
The pBrain constructs of >9.5 kb were difficult to work with, so later pBrain constructs were switched into a pBluescript SK+ backbone. These constructs were called pDiddy, due to their smaller size. The equivalent construct to pBrain-GFP-CHC(1-1639)KDP-CHC4 was made by first inserting a Sac II-Mfe I fragment from pBrain-GFP-CHC(1-1639)KDP-CHC4 into Sac II-EcoR I of pBluescript SK+, duplicated sites were removed by inserting complimentary annealed oligonucleotides (sense 5′-GGTAAGACTATC-3′ and antisense 5′-CCGGGATAGTCTTACCGC-3′) between Sac II and Xma I to give pDiddy-GFP-CHC(1-1639)KDP-CHC4. The knockdown construct to express GFP, pDiddy-GFP-CHC4 was made by inserting an Age I-Mlu I fragment from pEGFP-C1 into pDiddy-GFP-CHC(1-1639)KDP-CHC4. The control construct to express GFP, pDiddy-GFP-CHC1 was made by inserting a Sac II-Sac II fragment from pBrain-GFP-CHC1 into pDiddy-GFP-CHC4. These three pDiddy constructs gave results that were indistinguishable from their pBrain counterparts.
To make C1573S, an Xba I-Xba I piece containing the mutation was generated by the megaprimer method and inserted into pDiddy-GFP-CHC(1-1639)KDP-CHC4 to give pDiddy-GFP-CHC(1-1639)KDP(C1573S)-CHC4.
Any constructs that contained the sequence coding for CHC residues 60-66 (TCCAATTCGAAGACCAAT) were rendered knockdown-proof using silent mutations (to give TCCgATcaGgcGtCCtAT).
All constructs were verified by restriction digest and any that involved PCR were further verified by automated DNA sequencing (MRC Geneservice, UK). Details of primers used are given in Supplementary Table 2.
Cell biology
Human embryonic kidney HEK293 cells were cultured in DMEM containing 10% fetal bovine serum and 100 U/ml penicillin-streptomycin at 37°C and 5% CO2. Cells were plated at a density of 50,000/ml onto poly-l-lysine-coated cover slips or uncoated plastic Petri dishes for biochemistry. Transfection was carried out the next day using the calcium phosphate method (Bobanovic et al., 2002). Briefly, for 1 well of a 6-well plate (two cover slips) 150 μl CaCl2 (250 mM) and 3 μg DNA were mixed. 150 μl of 2X HBS was added dropwise, tube was shaken and the precipitates were left to form in the dark for 40 minutes. Precipitate was added to the cells and the media exchanged 8 hours later.
All cells were analysed 3 days post-transfection, when knockdown was maximal (Royle et al., 2005). For measurement of mitotic counts, cells were fixed in 3% PFA/4% sucrose for 10 minutes, nucleic acids were stained with H33342 (Sigma) and cover slips were mounted using ProLong (Molecular Probes). For transferrin uptake, cells were incubated in DMEM without serum for 15 minutes at 37°C and then in DMEM with 50 μg/ml transferrin-Alexa546 for 10 minutes, all at 37°C, 5% CO2 then fixed and mounted. For immunocytochemistry, cells were processed as previously described (Royle et al., 2002). Monoclonal antibodies against CHC (X22, Affinity BioReagents), α-tubulin (DM1A, Sigma) and CENP-B (kind gift from W.C. Earnshaw, University of Edinburgh, U.K.) and Alexa546- and Alexa647-conjugated secondary antibodies (Molecular Probes) were used.
Hydrodynamic methods
Sedimentation analysis was as previously described (Greger et al., 2003). Briefly, cells from 60 mm dishes were lysed in 250 μl lysis buffer (150 mM NaCl, 20 mM HEPES, pH 7.4, 2 mM EDTA, 5% glycerol, 0.6% CHAPS, 100 mM iodoacetamide, 1× protease inhibitor cocktail, 100 μg/ml PMSF) and extracted for 1 hr at 4°C and cleared by centrifugation at 16,000 × g for 30 minutes at 4°C. Soluble material (185 μl) was layered onto 15-40% or 10-35% glycerol gradients (glycerol w/v, 100 mM NaCl, 20 mM HEPES, pH 7.4, 2 mM EDTA, 0.1% Triton X-100). Gradients were poured using an automated pump and centrifuged in a SW-60 rotor at 45,000 r.p.m. for 18 hours at 4°C. 250 μl fractions were collected manually from the top. The refractive indices for samples from each gradient were verified to be linear using a refractometer (Zeiss). The 10-35% and 15-40% gradients ranged from 1.354 to 1.369 and 1.3565 to 1.371, respectively. Aliquots (25 μl) of each fraction were eluted with Laemmli sample buffer, subjected to SDS-PAGE (6% gel) and analysed by western blotting. Nitrocellulose membranes were probed with monoclonal anti-GFP (1:1000, JL-8, Clontech) and anti-mouse HRP-conjugated (1:1000) antibodies, signals were detected using ECL (Amersham). Films were scanned and densitometry performed using IPLab. Three molecular weight standards were used: bovine serum albumin 66 kDa, β-amylase from sweet potato 200 kDa and bovine thyroglobulin 669 kDa (MW-GF-1000, Sigma).
Imaging and data analysis
Confocal imaging was done using a BioRad Radiance 2000 and Nikon TE300 microscope with 60x (1.4 NA) or 100x (1.3 NA) oil immersion objectives. GFP and Alexa546 were excited using an Ar/Kr 488 nm and a He/Ne 543 nm laser, respectively. H33342 and Alexa647 were excited using 405 nm and 638 nm diodes, respectively. Excitation and collection of emission were performed separately and sequentially. Power output of the primary laser was checked regularly to ensure consistency (50 ± 1 mW; anode current 7.1 ± 0.2 A). For quantitative immunostaining experiments, identical laser power and acquisition settings were used. Images were captured to Lasersharp 5.0 software at a depth of 8-bit. Analysis of single-cell immunoreactivity from greyscale images was carried out essentially as described previously (Royle et al., 2005). Images were imported into IMAGEJ (NIH), the outline of the cell was manually drawn using the GFP channel and this ROI was transferred to the red channel. The mean pixel density was measured for X22 quantification or the image was thresholded and the number of transferrin-Alexa546 puncta was counted. Spindle recruitment was assayed by dividing the mean pixel density measured in a 1.08 μm2 ROI (8 × 8 pixel box) placed over the spindle (Fspindle) by that measured in a region outside the spindle (Fcytoplasm). Experiments to determine the mitotic index were done by counting the number of cells with mitotic figures as a proportion of the total number of GFP-positive cells within a 275 × 190 μm area. The number of GFP-positive metaphase-like cells that had misaligned chromosomes was counted. For mitotic index measurements between 256-2,733 cells per condition were counted and for frequency of misaligned chromosomes, between 63-290 metaphase-like cells were counted per construct. All experiments were performed a minimum of three times. Normally distributed data were analysed by ANOVA with Dunnet’s post-hoc test and binomial data were analysed by Fisher’s exact test or by Chi-square approximation using Instat (Graphpad). Values were compared to the GFP condition. Figures were assembled using Igor Pro (Wavemetrics), PyMOL (DeLano Scientific) and Adobe Photoshop.
Supplementary Material
Acknowledgements
We thank Ingo Greger for helpful advice and guidance on sedimentation analysis. We are very grateful to Phil Evans for generating the triskelion from the PDB co-ordinates 1XI4, which we used as the basis for Figure 1A. Finally, we thank Takahiro Nagase for the kind gift of human CHC cDNA. This work was supported by the HFSP and MRC.
References
- Bobanovic LK, Royle SJ, Murrell-Lagnado RD. P2X receptor trafficking in neurons is subunit specific. J. Neurosci. 2002;22:4814–4824. doi: 10.1523/JNEUROSCI.22-12-04814.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton DA. Spindle assembly in animal cells. Annu. Rev. Biochem. 2000;69:95–114. doi: 10.1146/annurev.biochem.69.1.95. [DOI] [PubMed] [Google Scholar]
- Fotin A, Cheng Y, Sliz P, Grigorieff N, Harrison SC, Kirchhausen T, Walz T. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432:573–579. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- Greger IH, Khatri L, Kong X, Ziff EB. AMPA receptortetramerization is mediated by Q/R editing. Neuron. 2003;40:763–774. doi: 10.1016/s0896-6273(03)00668-8. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T. Clathrin. Annu. Rev. Biochem. 2000;69:699–727. doi: 10.1146/annurev.biochem.69.1.699. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T, Harrison SC. Protein organization in clathrin trimers. Cell. 1981;23:755–761. doi: 10.1016/0092-8674(81)90439-6. [DOI] [PubMed] [Google Scholar]
- Liu SH, Wong ML, Craik CS, Brodsky FM. Regulation of clathrin assembly and trimerization defined using recombinant triskelion hubs. Cell. 1995;83:257–267. doi: 10.1016/0092-8674(95)90167-1. [DOI] [PubMed] [Google Scholar]
- Mack GJ, Compton DA. Analysis of mitotic microtubule-associated proteins using mass spectrometry identifies astrin, a spindle-associated protein. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14434–14439. doi: 10.1073/pnas.261371298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maro B, Johnson MH, Pickering SJ, Louvard D. Changes in the distribution of membranous organelles during mouse early development. J. Embryol. Exp. Morphol. 1985;90:287–309. [PubMed] [Google Scholar]
- Murphy JE, Keen JH. Recognition sites for clathrin-associated proteins AP-2 and AP-3 on clathrin triskelia. J. Biol. Chem. 1992;267:10850–10855. [PubMed] [Google Scholar]
- Nathke IS, Heuser J, Lupas A, Stock J, Turck CW, Brodsky FM. Folding and trimerization of clathrin subunits at the triskelion hub. Cell. 1992;68:899–910. doi: 10.1016/0092-8674(92)90033-9. [DOI] [PubMed] [Google Scholar]
- Okamoto CT, McKinney J, Jeng YY. Clathrin in mitotic spindles. Am. J. Physiol. Cell Physiol. 2000;279:C369–374. doi: 10.1152/ajpcell.2000.279.2.C369. [DOI] [PubMed] [Google Scholar]
- Ribbeck K, Groen AC, Santarella R, Bohnsack MT, Raemaekers T, Kocher T, Gentzel M, Gorlich D, Wilm M, Carmeliet G, et al. NuSAP, a Mitotic RanGTP Target That Stabilizes and Cross-links Microtubules. Mol. Biol. Cell. 2006 doi: 10.1091/mbc.E05-12-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle SJ, Bobanovic LK, Murrell-Lagnado RD. Identification of a non-canonical tyrosine-based endocytic motif in an ionotropic receptor. J. Biol. Chem. 2002;277:35378–35385. doi: 10.1074/jbc.M204844200. [DOI] [PubMed] [Google Scholar]
- Royle SJ, Bright NA, Lagnado L. Clathrin is required for the function of the mitotic spindle. Nature. 2005;434:1152–1157. doi: 10.1038/nature03502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey JM, Rogers GC, Sharp DJ. Mitosis, microtubules, and the matrix. J. Cell Biol. 2001;154:261–266. doi: 10.1083/jcb.200101097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, Vale RD, Mitchison TJ, Scholey JM. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J. Cell Biol. 1999;144:125–138. doi: 10.1083/jcb.144.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih W, Gallusser A, Kirchhausen T. A clathrin-binding site in the hinge of the beta 2 chain of mammalian AP-2 complexes. J. Biol. Chem. 1995;270:31083–31090. doi: 10.1074/jbc.270.52.31083. [DOI] [PubMed] [Google Scholar]
- Sutherland HG, Mumford GK, Newton K, Ford LV, Farrall R, Dellaire G, Caceres JF, Bickmore WA. Large-scale identification of mammalian proteins localized to nuclear sub-compartments. Hum. Mol. Genet. 2001;10:1995–2011. doi: 10.1093/hmg/10.18.1995. [DOI] [PubMed] [Google Scholar]
- Ungewickell E, Branton D. Assembly units of clathrin coats. Nature. 1981;289:420–422. doi: 10.1038/289420a0. [DOI] [PubMed] [Google Scholar]
- Wakeham DE, Chen CY, Greene B, Hwang PK, Brodsky FM. Clathrin self-assembly involves coordinated weak interactions favorable for cellular regulation. EMBO J. 2003;22:4980–4990. doi: 10.1093/emboj/cdg511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G. Membrane partitioning during cell division. Annu. Rev. Biochem. 1993;62:323–348. doi: 10.1146/annurev.bi.62.070193.001543. [DOI] [PubMed] [Google Scholar]
- Yao X, Abrieu A, Zheng Y, Sullivan KF, Cleveland DW. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2000;2:484–491. doi: 10.1038/35019518. [DOI] [PubMed] [Google Scholar]
- Ybe JA, Ruppel N, Mishra S, VanHaaften E. Contribution of cysteines to clathrin trimerization domain stability and mapping of light chain binding. Traffic. 2003;4:850–856. doi: 10.1046/j.1600-0854.2003.0139.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.