Figure 2.
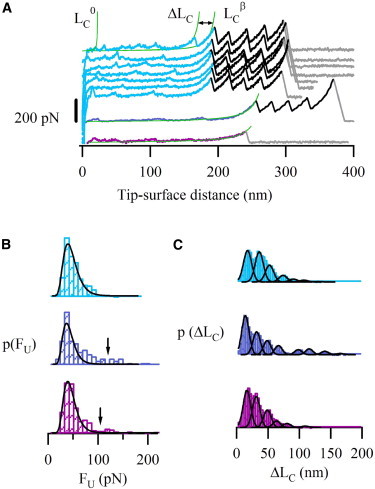
Single-molecule force spectroscopy of the ARM region and full-length β-catenin. (A) Force-extension recordings from the mechanical unfolding of the three constructs from Fig. 1C: ARMI27 (top, from LC0 to LCβ: ARM region; from LCβ to the last force peak: I27 modules), the βI27 (middle, from LC0 to LCβ: full-length β-catenin region; from LCβ to the last force peak: I27 modules) and β-catenin (bottom). Peaks were fitted using the worm-like chain model of polymer elasticity (smooth lines (26)), which allows to calculate contour lengths (LC0, ΔLC, and LCβ). The region under study in the three constructs unfolded in a similar way, with weak forces and variable ΔLC values (several traces from ARMI27 construct are shown as a representative example, Fig. S6). The I27 marker shows an ΔLC of 28 nm and a FU of around 200 pN, in accordance with previous reports (25). (B) FU histograms. Lines correspond to log-normal fittings (χ2 are 1.5 × 10−5, 8.5 × 10−5, and 2.3 × 10−5 for ARMI27, βI27 and β-catenin, respectively). Only 0.4% of the data points lie above 100 pN for ARMI27 (top), being 10% for βI27 (middle), and β-catenin (bottom) (containing unstructured terminal regions). This suggests that the ARM region is somehow stabilized by the outer regions. (C) The ΔLC histograms for the proteins show similar (though slightly shifted) multimodal distributions with the mean values close to the length of one or multiple ARM repeats (see details in the main text). ARMI27 (top), βI27 (middle), and β-catenin (bottom).
