Figure 2.
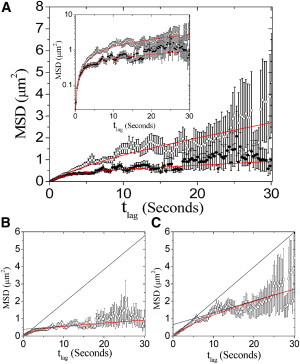
MSD versus time for prestin-SNAP-TMR. (A) Measured MSD against in untreated (solid circles) and cholesterol-depleted (open circles) cells. A total of 285 tracks were recorded for untreated cells (nine cells), and 269 tracks were recorded for cells treated with 10 mM MβCD (seven cells). Error bars are standard error of the mean. Increased variance at long result from decreased probability of observing long trajectories due to photobleaching of TMR. Both data sets are fit to the hop-diffusion model (Eq. 1, solid curve). Inset: Data are shown on a log scale to emphasize fit to early points. The resulting fitting parameters are (for untreated cells) , , and with ; and (for cells treated with MβCD) , , and with . Asymptotic behavior (black curves) of MSDhop for (B) untreated and (C) MβCD-treated groups is shown superimposed on data and fit (red curves). According to the hop-diffusion model, including averaging effects, and , where is a confinement offset seen as the y-intercept in B and C (28). Macroscopic diffusion constants : (B) and (C) .
