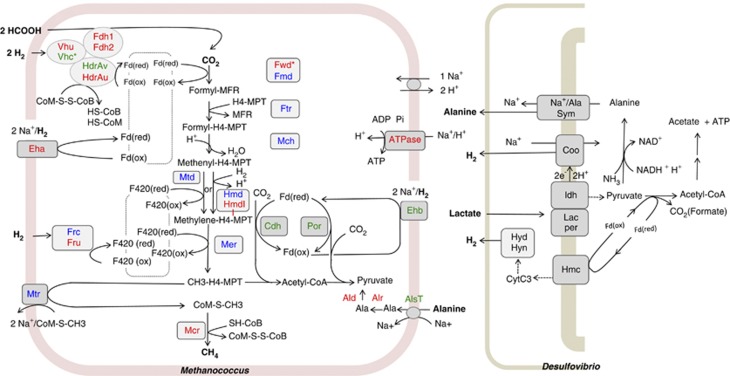
Figure 1.
Conceptual schematic of M. maripaludis and D. vulgaris syntrophic interaction, highlighting the central energy-generating and -consuming the pathways of the methanogen. Relative changes (Table 2) in transcript abundance during syntrophic growth are indicated by red (increase) and green (decrease) coloration. Blue coloration indicates no statistically significant change as specified in the Materials and methods section. Hydrogen-limited monocultures served as the control growth condition. Oxidation of formate to CO2 and H2 coupled with coenzyme F420-reduction (not depicted) is catalyzed by two alternative formate dehydrogenases, Fdh1 and Fdh2. One of two membrane-bound energy-conserving hydrogenases (Eha and Ehb) couple the chemiosmotic energy of ion gradients to H2 oxidation and ferredoxin reduction. Of these, Ehb generates the low potential electron carrier used for anabolism, whereas Eha is hypothesized to function primarily in the energy-generating methanogenesis pathway, generating low potential-reducing equivalents for the reduction of CO2 to formylmethanofuran (Major et al., 2010). Two different formylmethanofuran dehydrogenases catalyze this first step in methanogenesis, tungsten (Fwd) and molybdenum (Fmd) forms. Transfer of the formyl group from methanofuran to methanopterin by Ftr and subsequent elimination of H2O by Mch yields methenyl-H4-methanopterin. Two different enzymes can then reduce methenyl-H4-MPT to methylene-H4MPT, one (Mtd) using H2 as reductant and the other (Hmd) using reduced coenzyme F420. M. maripaludis has an Hmd paralog of unknown function (Mmp1716, HmdII) that may also function in this step (Hendrickson et al., 2004). Reduction of methylene-H4MPT by another F420-dependent reductase (Mer) yields methyl-H4MPT. The reduced coenzyme F420 required for the formation of methyl-H4MPT by these two steps is generated by one of two alternative F420-reducting hydrogenase (Fru and Frc). The final steps to methane production are catalyzed by a methyl transferase (Mtr) and a reductase (Mcr) coupled to two forms of a F420-nonreducing hydrogenase (Vhu and Vhc). The mixed disulfide (CoM-CoB) produced by reduction of methyl coenzyme M is then reduced by one of two forms of the heterodisulfide reductase determined by the composition of the HdrA subunit (HdrAU or HdrAV). Fdh/ Hdr/Vhu/ Fwd are reported to have protein–protein interactions (Costa et al., 2010). (Note: The interaction with Fwd could not be depicted here without compromising the clarity of the figure. Vhc is not part of this interaction). Other reactions include the transport of alanine (AlsT), and subsequent coversion to pyruvate via an alanine racemase (Alr) and dehydrogenase (Ald). Additional M. maripaludis proteins shown: pyruvate oxidoreductase, acetyl-CoA decarbonylase/synthase. The D. vulgaris metabolic pathway is based upon results as described in Walker et al., 2009 and is updated to include an unspecified sodium/alanine transporter (Na+/ala sym). Other D. vulgaris proteins shown: lactate permease (lac per), lactate deydrogenase (ldh), membrane-bound Coo hydrogenase (Coo), high-molecular weight cytochrome (Hmc), periplasmic hydrogenases (Hyd and Hyn), cytochrome c3 (Cyt c3) and oxidized and reduced ferredoxin (Fd).