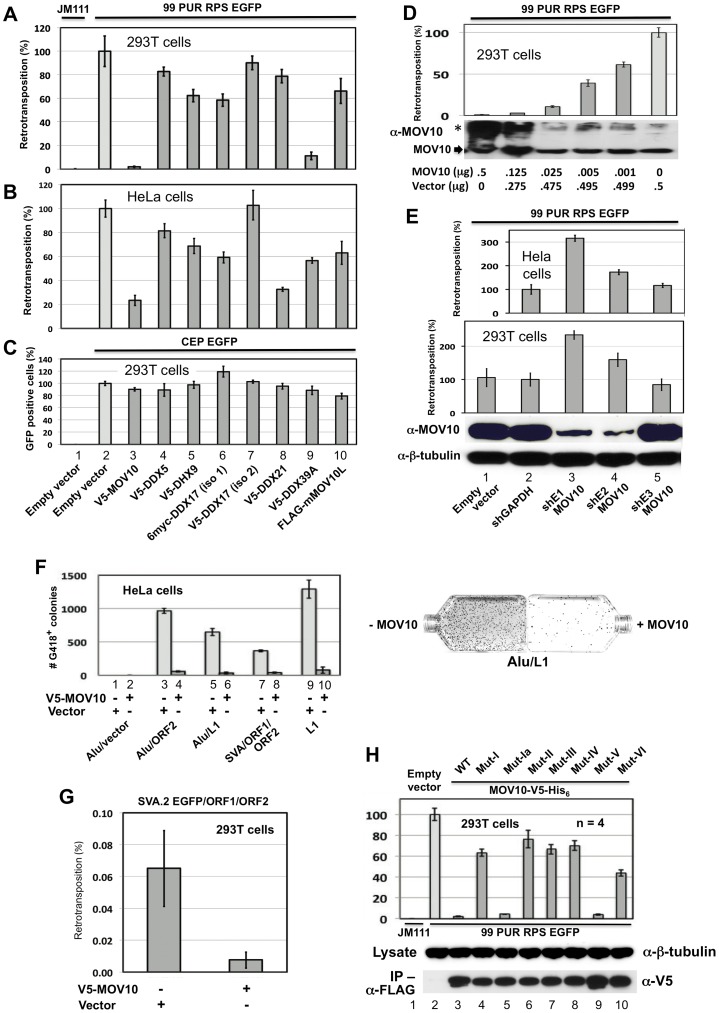
Figure 3. Evidence from cell culture retrotransposition assays that MOV10 inhibits insertion of non-LTR retrotransposons.
(A) The reporter construct 99 PUR RPS EGFP was cotransfected in 293T cells with empty vector (pcDNA3) or constructs expressing V5-MOV10 or other epitope-tagged L1 RNP-associated helicases. Constructs are numbered and named at the bottom of Panel C. Five days later, percentages of EGFP-positive cells (ie. cells with a retrotransposition event) were determined by flow cytometry. Each construct pair was tested in quadruplicate (n = 4), and results are normalized to pcDNA3 empty vector control (#2). An L1 mutant, 99 PUR JM111 EGFP (JM111), was used as negative control for retrotransposition and FACs gating (#1). V5-DDX17 (isoform (iso)2, #7), is generated from an abbreviated transcript variant spanning only residues 549 to 729 of the C-terminal region of full-length DDX17 (#6). (B) Same as (A) in HeLa-HA cells. (C) To assess transfection efficiency and cell toxicity, helicase constructs were cotransfected with CEP-EGFP and fluorescent cells were assayed 4 days later. Results are from 293T cells. (D) Expression of MOV10 inhibits retrotransposition in a dose-dependent manner. Decreasing microgram amounts of MOV10-expessing plasmid, mixed with empty vector to normalize DNA concentrations, were cotransfected with the L1-reporter construct, 99 PUR RPS EGFP, and assayed for retrotransposition at 5 days. Western blot analysis of cytoplasmic lysates (below) shows that even very small amounts of exogenous MOV10 protein can inhibit retrotransposition. High-molecular weight aggregates of overexpressed MOV10 protein are also visible on overexposed gels (marked by *). (E) Loss of endogenous MOV10 expression increases retrotransposition. Stable Hela and 293T cell lines were established by infection with lentiviral particles expressing shRNAs against MOV10, GAPDH or empty vector, followed by selection with puromycin. These cell lines were then tested for retrotransposition competency of 99 PUR RPS EGFP. Results are normalized to empty vector control. Bottom panels: Western blot showing that shE1 and shE2 decrease endogenous MOV10 protein levels by about 90 percent in 293T cells, and loading control blot showing β-tubulin. (F) Overexpression of MOV10 decreases Alu and SVA retrotransposition in HeLa-HA cells. As described in Dewannieux et al. [49], a Ya5 Alu is cloned in a plasmid containing the 7SL pol III enhancer and neoTET cassette interrupted by a Tetrahymena self-splicing intron. Upon transcription, the intron is spliced out. When this construct is co-expressed with L1 ORF2 alone (#3 and 4) or full-length L1-RP (#5 and 6), Alu RNAs are reverse transcribed along with the neo gene and integrated into the genome to confer neomycin resistance. Following 15 days of treatment with neomycin, resistant colonies were stained and counted. Either empty vector (#1, 3, 5, 7, and 9) or V5-MOV10 plasmid (#2, 4, 6, 8 and 10) was included in the reactions. Retrotransposition data for SVA.2 and L1-RP containing the mneoI antibiotic-selection cassette [47] are also shown (#7–10). Colony counts are not normalized. To the right are representative T75 flasks with Giemsa-stained Alu retrotransposition-positive colonies in the absence (left) or presence (right) of MOV10. (G) The SVA EGFP retrotransposition assay [28] was performed in 293T cells with SVA.2 EGFP cotransfected with constructs expressing L1 ORF1 and ORF2 separately, in the presence or absence of MOV10. (H) Mutations in helicase motifs I–VI impede anti-retrotransposition activity of MOV10. Top panel: MOV10-V5-His6 wild-type and mutant proteins [51] were tested for effect on retrotransposition of 99 PUR RPS EGFP. Results are normalized to empty vector control (#2). Middle panel: mutant and wild-type MOV10-V5-His6 proteins are expressed at similar levels in 293T cells. Bottom panel: mutant and wild-type MOV10-V5-HIS6 proteins all efficiently co-IP with L1 RNPs expressed from pc-L1-1FH.