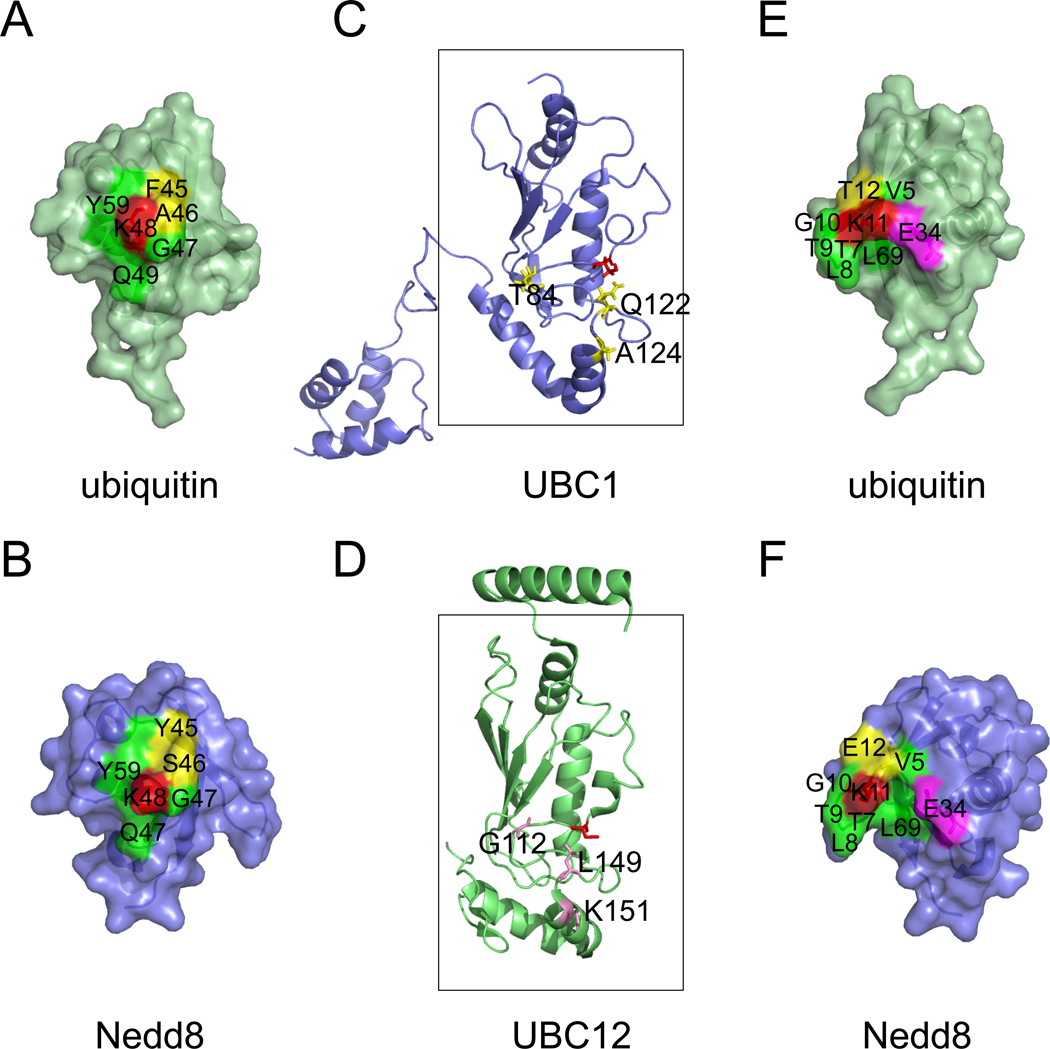
Figure 2. A structural comparison of the ubiquitin and the Nedd8 conjugation systems.
A, B, The environment surrounding Lys48 (red) of ubiquitin and the corresponding Lys in Nedd8 is not completely identical. Note that the yellow labeled residues are not conserved between ubiquitin and Nedd8. C, D, A comparison of UBC1 and UBC12. The boxes indicate the UBC core. Active site cysteines are shown in red. Residues critical for UBC1-dependent Lys48-linked polyubiquitination are shown in yellow. Note that the corresponding residues in UBC12 are different (labeled in magenta). E, F, The environment surrounding Lys11 of ubiquitin and the corresponding residue in Nedd8. Although most residues (labeled in green) in proximity to Lys11 (red) are conserved between ubiquitin and Nedd8, the position of the Glu34 side chain is distinct. In ubiquitin, Glu34 makes close contact with Lys11 to suppress its pKa for nucleophilic attack on an E2-ubiquitin thioester 62. Note that a 7.2A gap exists between Glu34 and Lys11 in Nedd8. The following structures were used for the analyses (Protein databank code (PBD) 1UBQ, ubiquitin 92; 1NDD, Nedd8 93; 1TTE, UBC1 94; 3O2U, UBC12 95). Note that a color version of the figure is available online.