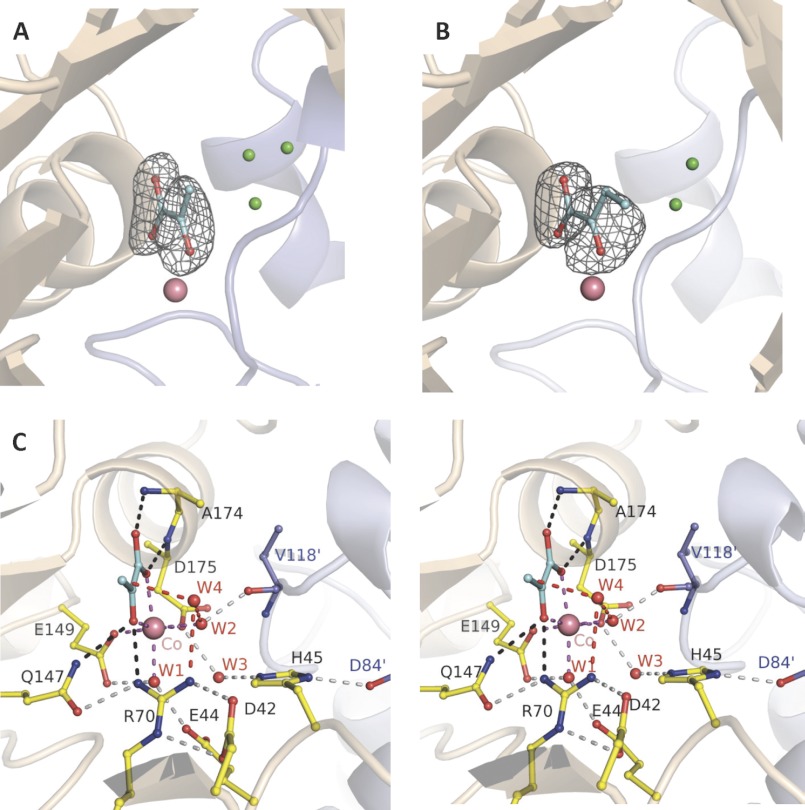
FIGURE 1.
Trapped 2-keto acids in active site of HpaI pyruvate aldolase. A, electron density of pyruvate bound in the active site of the native enzyme. B, electron density of ketobutyrate trapped in the active site of the native enzyme. C, stereoview showing active site interactions when pyruvate binds to HpaI aldolase. In A and B, water molecules occupying the aldehyde binding site (presented in Fig. 3) are shown in green. Magenta dashes depict coordination of Co2+ made by the two conserved water molecules (W1 and W2), pyruvate (blue), and residues Glu149 and Asp175. The backbone nitrogens of Ala174 and Glu175 direct the carboxylate oxygens of the pyruvate molecule, and Gln147 and Arg70 interact with the keto oxygen; these interactions are shown by black dashes. These interactions are used to specifically orient pyruvate in the active site. Hydrogen bonding interactions not directly contacting the metal ion or pyruvate (gray dots) organize remaining active site residues and water molecules. Interactions from Val118′ and Asp84′ of an adjacent contacting subunit (blue) complete the active site binding locus. The consistency in hydrogen bonding interactions made with W3 implies that Glu44 and Glu172 are hydrogen bond acceptors, whereas His45 is a hydrogen bond donor. Electron density encompassing the ligands was calculated from a simulated annealing Fo − Fc omit map and contoured at 3.5 σ.