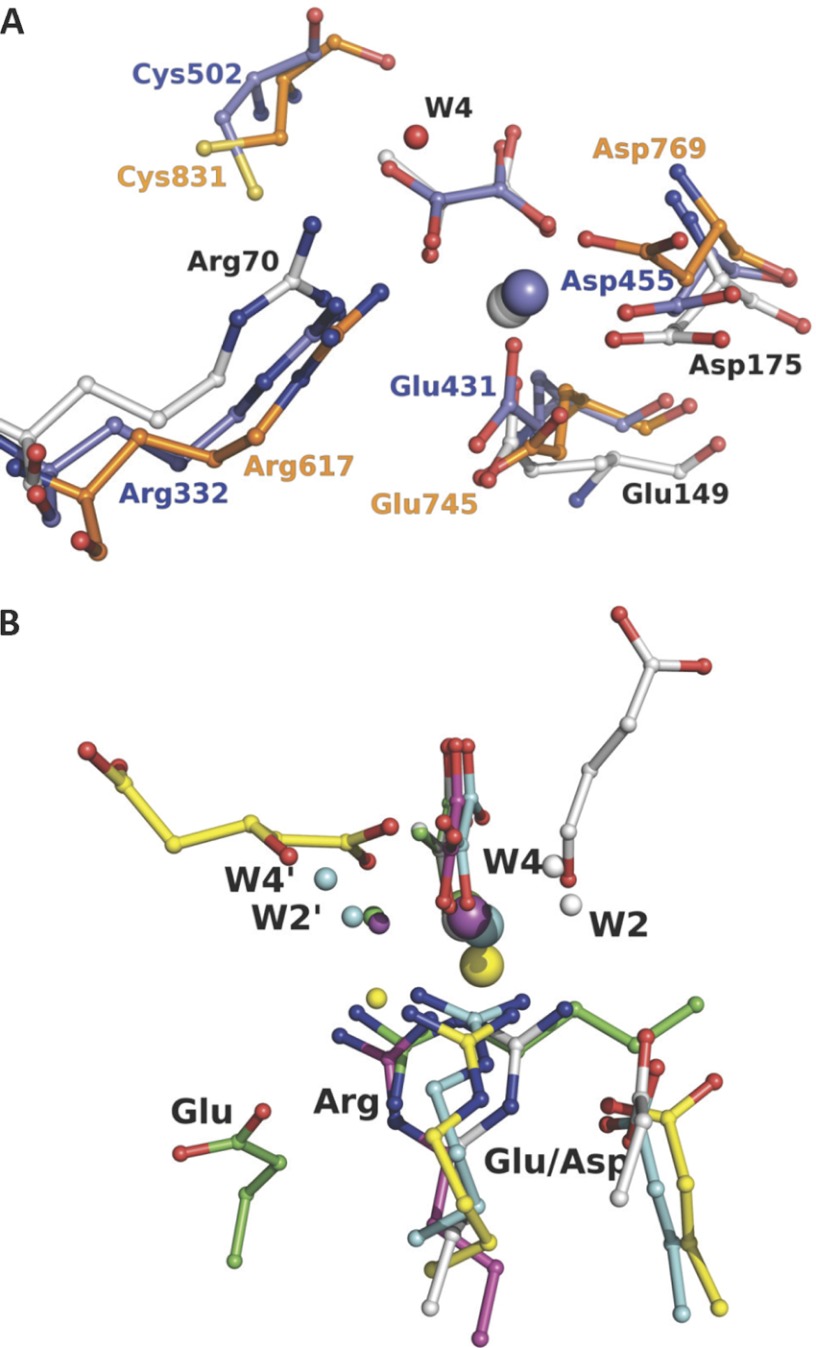
FIGURE 7.
Active site architecture comparison of evolutionary related enzymes and non-homologous pyruvate lyases. A shows superposition of the HpaI·pyruvate complex structure with the P-enolpyruvate binding domain of enzyme I for bacterial P-enolpyruvate-dependent carbohydrate:phosphotransferase systems (Protein Data Bank code 2BG5, residues 261–573) and pyruvate, phosphate dikinase (Protein Data Bank code 1KC7, residues 534–874). B shows superposition of HpaI pyruvate aldolase (white), human HMG-CoA lyase (Protein Data Bank code 2CW6) (yellow), DmpG aldolase (Protein Data Bank code 1NVM) (cyan), bacterial oxaloacetate decarboxylase (Protein Data Bank code 3B8I) (magenta), and HMG/CHA aldolase (Protein Data Bank code 3NOJ) (green). Structures are presented from the same perspective, taking the pyruvate analog and the metal ion (large sphere) as reference except for human HMG-CoA lyase as noted under “Material and Methods.” Succinic semialdehyde in HpaI aldolase is shown in white, and 3-hydroxyglutarate, a cleavage product of HMG-CoA lyase, is shown in yellow. For HpaI, the condensing aldehyde will approach from the distal side of the pyruvate, whereas for DmpG aldolase, HMG-CoA lyase, HMG/CHA aldolase, and oxaloacetate decarboxylase it will approach from the proximal side. Water molecules identified as W2′ and W4′, equivalent to W2 and W4, which are described as part of the catalytic machinery in HpaI aldolase, also show the same spatial disposition as would their respective condensing aldehydes. The guanidinium moieties of adjacent arginine residues cluster tightly (identified as Arg) and interact with their respective metal-binding ligand and water molecule identified as W4/W4′ where present. Water molecules within hydrogen bonding distance of W4/W4′ and bound to the metal ion are identified as W2/W2′. Note that the Arg70-Asp42 pair has a direct analog in HMG-CoA lyase (Arg41-Glu72), HMG/CHA aldolase (Arg123-Glu36), and DmpG aldolase (Arg17-Glu48), whereas only in oxaloacetate decarboxylase is the equivalent arginine residue, Arg159, not ion-paired with a carboxylate group of an amino acid. The carboxylate groups of the interacting aspartate or glutamate are identified (Asp or Glu).