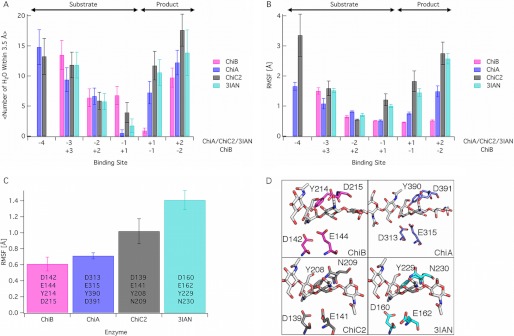
FIGURE 4.
MD simulations of two processive chitinases, ChiA and ChiB, and two nonprocessive chitinases, ChiC2 and 3IAN. All simulations were done on enzymes in which an oligomer of GlcNAc (GlcNAc5 in ChiB, GlcNAc6 in ChiA and 3IAN, and GlcNAc7 in ChiC2) had been docked to the −4 to +2 subsites of the reducing end enzymes and the +3 to −2 subsites of ChiB (see supplemental text for additional details). In panels A and B, only subsites appearing in all four enzymes are shown. The product and substrate subsite definitions are as shown above panel A. The panels show the average solvation of the ligand by binding site (A), the root mean square fluctuations (RMSF) of the ligand by the binding site (B), and the root mean square fluctuations of the tetrad of catalytic residues, listed for each enzyme (C). Error bars in panel A represent one standard deviation. Error bars in panels B and C were obtained with block averaging. D, the catalytic tetrad of each enzyme examined here with simulation with the residues labeled. These residues represent the set for analysis in panel C. The enzyme structures used for the simulation input are described in detail in the supplemental text.