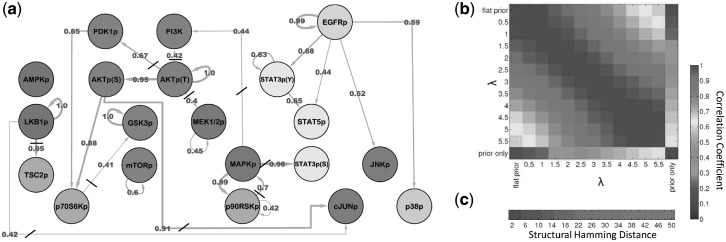
Fig. 3.
Data-driven signaling topology in the breast cancer cell line MDA-MB-468. Reverse-phase protein arrays were used to interrogate the phospho-proteins shown in (a), including key components of AKT, MAPK and STAT pathways, through time. DBNs were used to integrate the data with a network prior, derived from existing biology (Supplementary Fig. S3). Prior strength 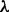 was set objectively using an empirical Bayes approach, resulting in
was set objectively using an empirical Bayes approach, resulting in 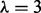 (see text and Supplementary Fig. S5). (a) Edges represent probabilistic relationships between proteins, through time. Edge labels indicate corresponding (posterior) probabilities (calculated exactly; edge thickness proportional to probability; all edges with probability
(see text and Supplementary Fig. S5). (a) Edges represent probabilistic relationships between proteins, through time. Edge labels indicate corresponding (posterior) probabilities (calculated exactly; edge thickness proportional to probability; all edges with probability  0.4 shown; strikethroughs ‘/’ indicate edges not expected under the network prior; Supplementary Figure S6 shows heatmap of all 400 posterior edge probabilities; list of proteins (and antibodies) given in Supplementary Table S1). (b) Sensitivity to prior strength. Results were compared over a range of values of
0.4 shown; strikethroughs ‘/’ indicate edges not expected under the network prior; Supplementary Figure S6 shows heatmap of all 400 posterior edge probabilities; list of proteins (and antibodies) given in Supplementary Table S1). (b) Sensitivity to prior strength. Results were compared over a range of values of 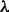 plus the flat prior (i.e.
plus the flat prior (i.e. 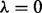 ) and prior only. Heatmap shows Pearson correlations between edge probabilities for pairs of prior regimes. Results were not sensitive to precise value of
) and prior only. Heatmap shows Pearson correlations between edge probabilities for pairs of prior regimes. Results were not sensitive to precise value of 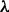 and differed markedly from prior alone. (c) Sensitivity to prior graph. The prior graph was perturbed and results obtained compared with those reported (see Supplementary Text for details). Correlation (as in (b)) is shown as a function of number of edge changes in the prior graph (‘Structural Hamming Distance’, see Supplementary Fig. S7 for error bars)
and differed markedly from prior alone. (c) Sensitivity to prior graph. The prior graph was perturbed and results obtained compared with those reported (see Supplementary Text for details). Correlation (as in (b)) is shown as a function of number of edge changes in the prior graph (‘Structural Hamming Distance’, see Supplementary Fig. S7 for error bars)