Fig. 1.
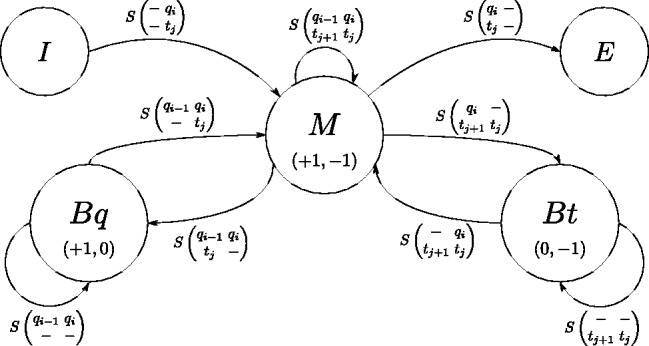
The flow in the three-state model. This state model has been (developed for and) proven useful before in the pairwise alignment of amino acid sequences using doublets, hereby taking into account correlation of neighboring residues (Akbasli, 2008). Here, the dashes indicate bulges or asymmetric internal loops, but are equivalent to gaps when the state model is applied to sequence alignments. States are represented by circles, transitions by connecting arcs. The number of pairs in the circles indicate the index increments to reach that state, e.g. for the Bq-state (bulge in query) only the q(uery) index is incremented, thus ‘(+1,0)’, while the M-state is reached by (mis)matching two residues, so both indices are updated. In a DP matrix, this corresponds to moving diagonally for transitioning into the M-state, and horizontally/vertically for the B-states. Indices along the t(arget) sequence are decremented as the two interacting RNA strands run in opposite directions. See recursion in the text for further description
