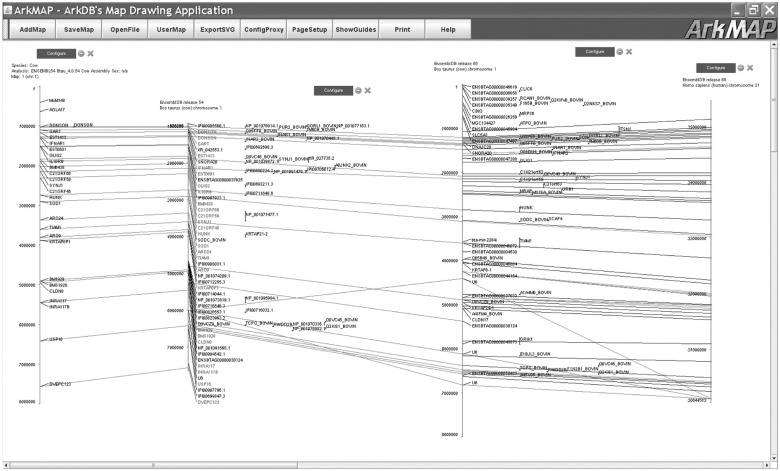
Fig. 6.
The ArkMAP application uses JEnsembl for retrieving maps and homologies from Ensembl datasources. ArkMAP can be used to draw genetic maps loaded from ArkDB, Ensembl or local datasources. Here the first 8 Mb of a bovine ePCR map has been loaded from ArkDB, where Ark Markers have been mapped on the Btau4 assembly. The JEnsembl API was then used to retrieve and align the cognate gene-annotated chromosome 1 assembly from Ensembl release 54. JEnsembl was then used to retrieve a more recent (release 66) gene annotated assembly which is aligned to the old assembly. Finally, JEnsembl was used to search for human gene homologies with the bovine genes in this region, and the region of conserved synteny on human chromosome 21 aligned with the bovine chromosome (with colour-coded homology relationships)