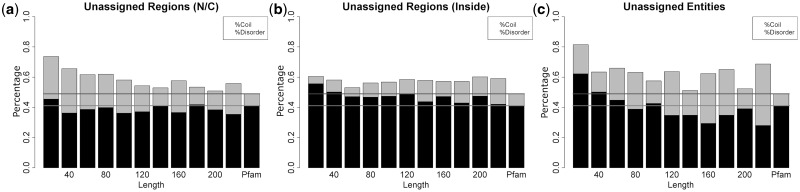
Fig. 4.
Secondary structure of the unassigned regions/sequences and Pfam assignments in PDBfam. Disorder means the residues do not have coordinates in the PDB file, whereas coil includes all residues that are not α-helix or β-strand. The y-axis is the number of coil plus disordered residues divided by the total unassigned residues at that length range. A bar is 20 amino acids long, i.e. 1–20, 21–40, 41–60, etc. The last bar in each figure shows the values for Pfam assignments. The next to last column in each figure shows the values for all regions with length >200. The light grey horizontal line is the rate of coil in the Pfam assigned regions, and the dark grey horizontal line is the rate of coil plus disorder in the Pfam assignments