Abstract
Genome editing is a powerful technique that can be used to elucidate gene function and the genetic basis of disease. Traditional gene editing methods such as chemical-based mutagenesis or random integration of DNA sequences confer indiscriminate genetic changes in an overall inefficient manner and require incorporation of undesirable synthetic sequences or use of aberrant culture conditions, potentially confusing biological study. By contrast, transient ZFN expression in a cell can facilitate precise, heritable gene editing in a highly efficient manner without the need for administration of chemicals or integration of synthetic transgenes.
Zinc finger nucleases (ZFNs) are enzymes which bind and cut distinct sequences of double-stranded DNA (dsDNA). A functional CompoZr ZFN unit consists of two individual monomeric proteins that bind a DNA "half-site" of approximately 15-18 nucleotides (see Figure 1). When two ZFN monomers "home" to their adjacent target sites the DNA-cleavage domains dimerize and create a double-strand break (DSB) in the DNA.1 Introduction of ZFN-mediated DSBs in the genome lays a foundation for highly efficient genome editing. Imperfect repair of DSBs in a cell via the non-homologous end-joining (NHEJ) DNA repair pathway can result in small insertions and deletions (indels). Creation of indels within the gene coding sequence of a cell can result in frameshift and subsequent functional knockout of a gene locus at high efficiency.2 While this protocol describes the use of ZFNs to create a gene knockout, integration of transgenes may also be conducted via homology-directed repair at the ZFN cut site.
The CompoZr Custom ZFN Service represents a systematic, comprehensive, and well-characterized approach to targeted gene editing for the scientific community with ZFN technology. Sigma scientists work closely with investigators to 1) perform due diligence analysis including analysis of relevant gene structure, biology, and model system pursuant to the project goals, 2) apply this knowledge to develop a sound targeting strategy, 3) then design, build, and functionally validate ZFNs for activity in a relevant cell line. The investigator receives positive control genomic DNA and primers, and ready-to-use ZFN reagents supplied in both plasmid DNA and in-vitro transcribed mRNA format. These reagents may then be delivered for transient expression in the investigator’s cell line or cell type of choice. Samples are then tested for gene editing at the locus of interest by standard molecular biology techniques including PCR amplification, enzymatic digest, and electrophoresis. After positive signal for gene editing is detected in the initial population, cells are single-cell cloned and genotyped for identification of mutant clones/alleles.
Keywords: Genetics, Issue 64, Molecular Biology, Zinc Finger Nuclease, Genome Engineering, Genomic Editing, Gene Modification, Gene Knockout, Gene Integration, non-homologous end joining, homologous recombination, targeted genome editing
Protocol
1. Custom ZFN Design Process
- To expedite the ZFN design process, an investigator should provide:
- Information on the target gene such as the gene name, species, and gene annotation/identification number.
- A clearly stated over-all objective of the experiment (e.g., Knockout or Knockin)
- Any specific factors of the gene target which include atypical gene structure, special biology implicated in the genetic locus, or any homologous regions elsewhere in the genome.
- The specific range of DNA sequence for the zinc finger nucleases to target.
The Sigma bioinformatics team conducts in silico ZFN design to develop initial zinc finger target sites.
Initial ZFN target sites are provided to a ZFN scientific technical consultant. For more technically complex projects, this Sigma scientist works with the investigators to develop the project strategy.
The Sigma bioinformatics team conducts an analysis with ZFN algorithms to generate in silico ZFN designs. This includes a whole genome search for off-target sites, repeat masking, and SNP analysis for each potential ZFN target site.
Bioinformatics provides the top ZFN target sites to the investigator for review. Upon approval of the potential ZFN target sites, this information is provided to the ZFN Production team to begin the ZFN manufacturing process.
2. Custom ZFN Production
The Sigma archive of zinc finger modules is used to assemble the approved ZFN designs.
Each ZFN design is assembled and sequence verified on a high throughput cloning platform.
Once production of all ZFN designs is complete every ZFN is sent to validation.
3. Validation of Custom ZFNs
ZFN projects in human, mouse, rat or CHO species are validated in well characterized cell lines for each species, a yeast based assay is used for other organisms or cell types.
ZFN constructs are delivered into the appropriate cell line by nucleofection and the ZFNs are expressed.
DNA is harvested from the pool of ZFN transfected cells and the region of interest is PCR amplified.
The Cel-1 assay is then conducted on the PCR product. Gel electrophoresis is run on the Cel-1 assay to validated ZFN activity.
The highest activity ZFN design as confirmed by the Cel-1 assay is then provided to the customer. ZFNs are delivered in mRNA and plasmid DNA along with PCR primers for the Cel-1 assay and genomic DNA as a positive control.
4. Delivery of Validated ZFNs by Nucleofection
Seed the cells at a density of 2x105 cells/ml the day before nucleofection.
On the day of nucleofection, take out Cell Line Nucleofector Kit V and let warm to room temperature.
Add the supplement to the Nucleofection Solution V according to the manufacturer's protocol.
Count the cells. Cell density should be between 2.5-5x105 cells/ml.
Fill a 6-well plate with 2ml of medium in each well and pre-warm in a CO2 incubator at 37 °C for at least 20 minutes prior to nucleofection.
Centrifuge 2x106 cells per transfection (8x106 total) at 200xg for 5 minutes.
Wash cells twice with 20 ml of Hank's Balanced Salt Solution (HBSS).
Prepare experimental tubes: (see table in Custom ZFN technical bulletin)
Remove the 6-well plate containing media from step (6.5) from incubator.
Resuspend cells in 400 μl (100 μl/reaction) of Nucleofection Solution V.
One reaction at a time, add 100 μl of cells to each DNA or mRNA-containing tube. Transfer the mixture to a 2 mm electroporation cuvette and nucleofect on a Nucleofector with the appropriate program.
Immediately after nucleofection of each sample, use a transfer pipette to add ~500 μl of prewarmed medium from the 6-well plate in step (6.9) to the cuvette. Then, carefully transfer cells from the cuvette to the remaining prewarmed medium in the 6-well plate.
Finish all reactions and return the 6-well plate to the CO2 incubator at 37 °C.
5. Harvesting Genomic DNA after Delivery of ZFNs
6-well plate – do not harvest all of the pooled cells. It is important to maintain the culture in order to have cells to single cell dilution clone after you confirm that you have Cel-1 digestion products. One to three days after nucleofection collect the cells to prepare chromosomal DNA using the GenElute Mammalian Genomic DNA Miniprep Kit.
6. Cel-1 Assay
PCR amplify the genomic DNA from the ZFN transfected samples and the positive control genomic DNA provided in the kit, using the supplied primers. The PCR reaction is set up with the following conditions: (see table in Custom ZFN technical bulletin).
To generate heteroduplexes from the PCR homoduplexes take 10 μl of PCR reaction from each ZFN treated sample plus the control and use the following program on a thermocycler: 95 °C, 10 minutes 95 °C to 85 °C, -2 °C/second 85 °C to 25 °C, -0.1 °C/second 4 °C, indefinitely
Add 1 μl of enhancer and 1 μl of Nuclease S (from Transgenomic Catalog Number 706025) to each reaction and incubate at 42 °C for 20-40 minutes.
Run the digestions on a 10% PAGE-TBE gel with proper markers, such as DirectLoad WideRange DNA Ladder (Catalog Number D7058) (see results in Custom ZFN technical bulletin).
Once positive signal has been detected in ZFN-treated populations by cel-I assay, proceed to clonal isolation and characterization.
7. Clonal Isolation and Characterization
Single-cell clone the ZFN-treated samples by standard methods including FACS and dilution cloning.
Allow the clones to expand sufficiently, then split some proportion of cells for genomic DNA harvest, freezing back the remainder as banked material.
Amplify genomic DNA from the clones using the Cel-I primers and analyze the amplicon by Cel-I assay (with and without WT DNA spiked in), or alternatively use this amplicon to proceed directly to genotyping.
Genotype candidate clones identified as having edited alleles by the preferred method.
8. Representative Results
An example of the entire CompoZr ZFN workflow is presented in Figure 2. ZFNs are delivered to cells where they bind and cleave the appropriate DNA sequence which creates a double-stranded break (DSB). The natural repair process, non-homologous end joining (NHEJ), repairs the DSB. In some instances aberrant NHEJ results in deletion, insertion or substitution of nucleotides. PCR amplification of harvested genomic DNA results in heteroduplex formation between wild type and modified amplicons after the denaturation/annealing step of the PCR reaction. Addition of the Cel-1 enzyme results in cleavage of any heteroduplex molecules. Cel-1 results are resolved by PAGE analysis to confirm ZFN cleavage. Figure 3 provides a schematic of the Cel-1 assay and the expected results.
Representative Cel-1 results are contained in Figures 4 and 5. Figure 4 contains results from a very active pair of ZFNs (21%) in K562 cells; Figure 5 contains results from a less active pair (2.4%) of ZFNs in K562 cells. ZFN activity is confirmed in both figures by the presence of two PCR fragments below the parental PCR fragment.
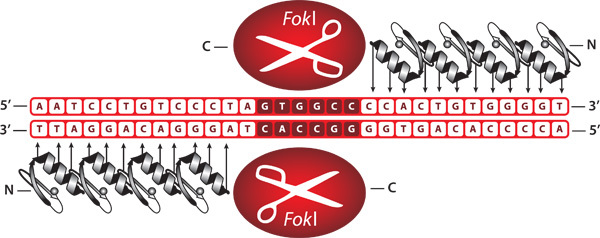 Figure 1. Representation of bound zinc finger nucleases. ZFNs are engineered proteins comprised of a zinc finger DNA-binding domain fused to the cleavage domain of the FokI restriction endonuclease. When bound as a heterodimer, ZFNs create a double-stranded break at a user specified DNA sequence.
Figure 1. Representation of bound zinc finger nucleases. ZFNs are engineered proteins comprised of a zinc finger DNA-binding domain fused to the cleavage domain of the FokI restriction endonuclease. When bound as a heterodimer, ZFNs create a double-stranded break at a user specified DNA sequence.
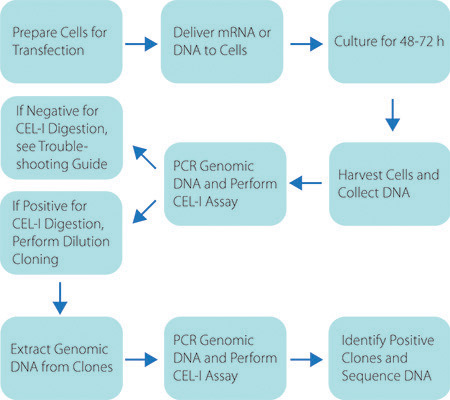 Figure 2. Schematic of the Custom ZFN Service Workflow. Graphic representation of the workflow to generate a genetically modified cell line using CompoZr ZFNs.
Figure 2. Schematic of the Custom ZFN Service Workflow. Graphic representation of the workflow to generate a genetically modified cell line using CompoZr ZFNs.
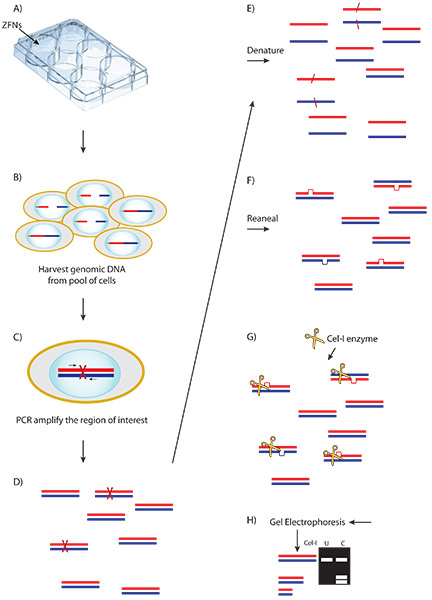 Figure 3. Schematic of the Cel-1 assay and results. A) ZFN plasmid or mRNA is delivered to cells. B) Expressed ZFNs bind and cut their target sequence creating a double stranded break (DSB) in a portion of cells. C) Aberrant repair of some DSBs by non-homologous end joining results in nucleotide insertion or deletion. D) Genomic DNA is harvested from the transfected pool of cells and amplified at the locus of interest. E-F) PCR product is denatured and re-annealed creating heteroduplex formation between wild type and modified amplicons. G) The Cel-1 mismatch endonuclease assay results in cleavage of heteroduplex molecules. H) Cel-1 enzyme digests are resolved by PAGE. The observed ratio of cleavage product to parenteral band, determined by ImageJ software, indicates the fraction cut and efficiency of the ZFNs.
Figure 3. Schematic of the Cel-1 assay and results. A) ZFN plasmid or mRNA is delivered to cells. B) Expressed ZFNs bind and cut their target sequence creating a double stranded break (DSB) in a portion of cells. C) Aberrant repair of some DSBs by non-homologous end joining results in nucleotide insertion or deletion. D) Genomic DNA is harvested from the transfected pool of cells and amplified at the locus of interest. E-F) PCR product is denatured and re-annealed creating heteroduplex formation between wild type and modified amplicons. G) The Cel-1 mismatch endonuclease assay results in cleavage of heteroduplex molecules. H) Cel-1 enzyme digests are resolved by PAGE. The observed ratio of cleavage product to parenteral band, determined by ImageJ software, indicates the fraction cut and efficiency of the ZFNs.
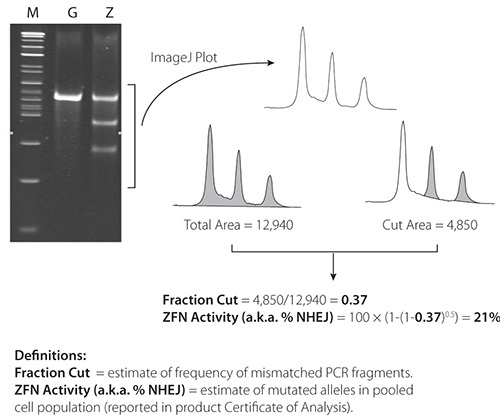 Figure 4. Cel-1 results from 21% active ZFNs. ImageJ software is available at: http://rsbweb.nih.gov/ij/.
Figure 4. Cel-1 results from 21% active ZFNs. ImageJ software is available at: http://rsbweb.nih.gov/ij/.
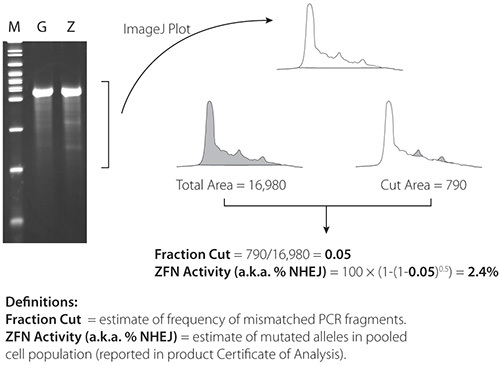 Figure 5. Cel-1 results from 2.4% active ZFNs. ImageJ software is available at: http://rsbweb.nih.gov/ij/.
Figure 5. Cel-1 results from 2.4% active ZFNs. ImageJ software is available at: http://rsbweb.nih.gov/ij/.
Discussion
Once ZFN-modified cells are isolated they have permanent and heritable DNA modifications. This results in the ability to generate stably modified cell lines or breed animals with desired genetic modifications to conduct research. CompoZr Custom ZFNs provide a robust method for genome editing in a wide variety of cell types. Publication of successful genome editing with ZFNs includes but isn't limited to human cell lines, mouse, rat, zebrafish, frog, porcine and CHO research models.3,4,5,6,7,8,9 The ability to create a double-stranded break at the desired target site in the specified cell type is assured when the ZFNs are properly delivered into a cell. It's also possible to perform sequential zinc finger modifications to a cell in order to have more than one genetic modification within a cell.10 The ability to select a specific target sequence and only modify that particular locus is a primary cause for the ability to create multiple modifications.
The CompoZr Custom ZFN certificate of analysis is generated with the very reagents provided in the kit thus assuring that all components of the kit have been thoroughly validated for customer success. Critical steps of the workflow from beginning to end are presented below:
Cell cloning
The ability to isolate and expand a given cell line should be tested before beginning any ZFN experiments. Single cells should be isolated and expanded to ensure that a clonally-derived population is possible. Some cell lines are recalcitrant in this matter, and tricks such as enrichment for edited clones, or culture with conditioned media can help to overcome these challenges.
Delivery efficiency
Optimization of delivery efficiency is a must before delivery of the ZFNs. Many methods may be used including lipid-based transfection, electroporation, and nucleofection. The ideal delivery method consists of one that affords the most optimal blend of delivery efficiency and cell survival. Quantitating these efficiencies may be estimated by delivering a GFP control plasmid and quantitating by visual inspection or FACS 24-48hrs post-delivery.
Cel-I assay
It is imperative that Cel-I assay is performed in parallel with the appropriate controls to ensure that PCR, digestion, and electrophoresis parameters will not interfere with the interpretation of ZFN experiment. Each kit includes control genomic DNA that is used to create the Cel-I image in the certificate of analysis. Amplification from the control DNA with the provided primers followed by Cel-I assay will allow the user to compare the results with that provided in the Certificate of Analysis (CofA) image, and isolate any potential inefficiencies in this aspect of an experiment. An appropriate negative control such as a mock, or GFP treated cell sample should also be included to allow elucidation of any non-specific cleavage products.
Qualitative/Quantitative analysis of single-cell clones
After ZFN treatment and single cell cloning, populations of cells may be screened for editing at the locus of interest by a number of methods. Cel-I assay may be conducted on genomic DNA from clones for the purpose of identifying candidate clones for further genotyping. It is important to spike WT PCR amplicon into the sample amplicons at a 1:1 ratio before performing the Cel-I digest as homozygous mutant clones will contain homogeneous molecules, and thus convey a negative Cel-I assay result. An alternative method for screening these candidate clones is to design one PCR primer landing directly on the ZFN target site. Used in conjunction with one of the control primers, a negative assay result indicates loss of the WT sequence. A heterozygous clone containing at least one WT allele will yield PCR signal, but may be segregated from fully WT clones by performing the PCR in the context of SYBR qPCR.
Once candidate clones have been identified by the above methods, they may be genotyped by standard pyrosequencing, deep sequencing, or other sequencing methods. For all sequencing methods, an amplicon created from the clonal genomic DNA should be generated. For traditional pyrosequencing alleles may be segregated by TA-cloning the PCR product, and sequencing the individual colonies. For methods such as deep sequencing this step is not necessary.
Homology Directed Repair (HDR)
This protocol provides the method for ZFN mediated gene knockout via NHEJ. Integration of transgenes may also be conducted by introduction of a donor plasmid with homology arms that flank a ZFN cut site.11 In this scenario, the ZFN created double-stranded break is repaired by HDR instead of NHEJ. HDR directed integration of transgenes may be confirmed using similar methods to the procedures provided throughout this protocol.
Troubleshooting
Delivery of ZFNs
ZFN delivery and expression- delivery may be controlled for by delivery of a GFP or other such plasmid for visualization and/or quantitation. Low expression due to promoter incompatibility with the cell type of interest may be overcome by delivery of ZFNs in mRNA format. Additionally, the cold shock method may be utilized to further increase the efficacy of ZFNs in cells.12 If mRNA is used it is imperative that cells are washed prior to delivery to ensure all serum-derived Rnases have been removed. It is also prudent to thaw the mRNA on ice, and add the it to the cells at the very last moment to minimize any chance for degradation.
Cel-I assay
The foundation of the Cel-I assay is specific and ample amplification of the locus of interest. If non-specific amplification products are observed use standard PCR troubleshooting procedures to increase specificity such as optimization of template amount, primer concentration, and cycling parameters. Perform PAGE analysis as opposed to standard agarose electrophoresis as the latter method does not provide adequate resolution and sensitivity. Cel-I digest conditions may also be optimized if no digested product or excessive smearing is observed. Titration of the amount of Cel-1 nuclease and/or length of digestion time can be titrated to improve these aspects.
Disclosures
Researchers can provide ZFN modified cells or animals to collaborators under the standard Research License Agreement. In this circumstance Sigma asks that a material transfer agreement is put in place between Sigma, the ZFN customer and their collaborator. The research license may be viewed at: http://www.sigma.com/zfn.
If there is a potential for a commercial application using ZFN modified cells Sigma then asks that a Commercial license is put in place.
Production and Free Access to this article is sponsored by Sigma, Inc.
References
- Kim YG. Hybrid restriction enzymes: zinc finger fusions to FokI cleavage domain. Proc. Natl. Acad. Sci. U.S.A. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Moehle EA. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc. Natl. Acad. Sci. U.S.A. 2007;104:3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc. Natl. Acad. Sci. U.S.A. 2010;107:15022. doi: 10.1073/pnas.1009424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts AM. Knockout rats via microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JJ. Efficient targeted gene disruption in the soma and germ line of the frog Xenopus tropicalis using engineered zinc-finger nucleases. Proc. Natl. Acad. Sci. U.S.A. 2011. Forthcoming. [DOI] [PMC free article] [PubMed]
- Wanatabe M. Knockout of exogenous EGFP gene in porcine somatic cells using zinc-finger nucleases. Biochem. Biophys. Res. Commun. 2010;402:14–18. doi: 10.1016/j.bbrc.2010.09.092. [DOI] [PubMed] [Google Scholar]
- Santiago Y. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui PQ. Generation of a triple-gene knockout mammalian cell line using engineered zinc-finger nucleases. Biotechnol. Bioeng. 2010;106:97–105. doi: 10.1002/bit.22654. [DOI] [PubMed] [Google Scholar]
- Moehle EA. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc. Natl. Acad. Sci. U.S.A. 2007;104:3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y. Transient cold shock enhances zinc-finger nuclease mediated gene disruption. Nat. Methods. 2010;7:459–460. doi: 10.1038/nmeth.1456. [DOI] [PubMed] [Google Scholar]


