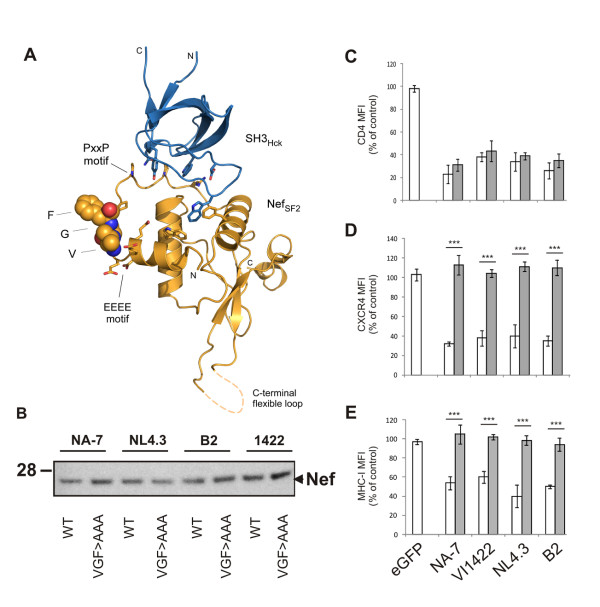
Figure 3.
The VGF linker region is critical for downregulation of CXCR4 and MHC-I by HIV-1 Nef. To evaluate the functional importance, VGF→AAA mutants of several Nef proteins were expressed and evaluated for their effect on cell surface receptor expression. (A) Display of the VGF motif in the structure of NefSF2 (yellow ribbon) bound to an Hck-SH3 domain (blue ribbon). VGF residues, which are deleted in Nef allele O8, are indicated as space-filling model. Residues of the preceding acidic cluster and the following PxxP motif are shown as stick representation. The structure is based on PBD accession number 3REA [23]. (B) WT or VGF→AAA Nef-IRES-eGFP transduced Jurkat CD4-CCR5 cells were sorted, lysed and analyzed for Nef expression. The figure shows a Western blot of whole cell lysates stained with sheep-anti-Nef antiserum. The marker line indicates position of a 28 kDa standard. Loading control staining (actin) was similar over all samples (not shown). (C-E) Relative CD4, MHC-I, CXCR4 surface levels measured by flow cytometry on Jurkat CD4-CCR5 cells transduced with retroviral vectors expressing either eGFP alone or together with the indicated nef alleles and mutants. To determine relative surface levels, the MFI of transduced cells (eGFP+) was divided by the MFI of untransduced cells (eGFP-) in the same culture. Data represent the mean relative expression and standard deviations obtained from 3 independent experiments, wild-type allele (open columns) next to the corresponding VGF→AAA mutants (grey shaded columns). *** over a bar indicates a statistically significant difference between the VGF→AAA mutant compared to the wild-type allele (P < 0.001).