Figure 1.
OGA Binds Different O-GlcNAc Glycopeptides with Similar Conformation but Different Affinities
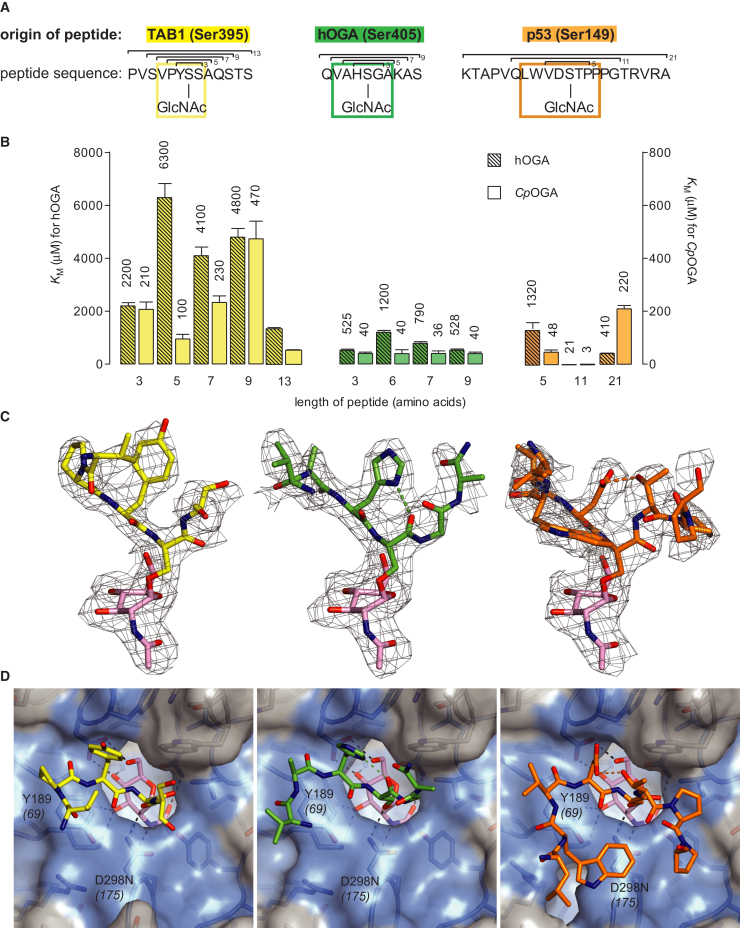
(A) Origin and sequence of O-GlcNAc peptides. The sequence of the longest peptides used is given, with residues observed in the crystal structure highlighted by colored boxes, and the shorter peptides indicated by brackets.
(B) Km of glycopeptide substrates for hOGA (displayed by filled bars [values for the three shortest hOGA- and TAB1-derived peptides reproduced from Schimpl et al., 2010]) and CpOGA (unfilled bars, right y axis) as determined by substrate competition assay with error bars representing the error of curve-fit (see Experimental Procedures for details).
(C) Conformation of O-GlcNAc peptides bound in the active site of CpOGA as determined by X-ray crystallography. Peptides are shown as sticks with colored carbons (TAB1 peptide, yellow; hOGA peptide, green; p53 peptide, orange), with the GlcNAc sugar highlighted by pink carbons. Unbiased |Fo|−|Fc|,Φcalc electron density (i.e., before addition of any glycopeptide model) is shown in gray (contoured at 2.25 σ). Intramolecular hydrogen bonds are shown by dashed lines.
(D) O-GlcNAc peptides (sticks) in complex with CpOGA D298N (surface representation). Sequence conservation between hOGA and CpOGA is indicated by blue shading of identical residues on the molecular surface. The catalytic acid Asp298 (mutated to Asn) and the conserved Tyr198 are labeled, with corresponding residue numbers for hOGA given in brackets.
See also Figures S1 and S2.