Abstract
We propose that haplotyped loci with high heterozygosity can be useful in human identification, especially within families, if recombination is very low among the sites. Three or more SNPs extending over small molecular intervals (<10 KB) can be identified in the human genome to define miniature haplotypes with moderate levels of linkage disequilibrium. Properly selected, these mini-haplotypes (or minihaps) consist of multiple haplotype lineages (alleles) that have evolved from the ancestral human haplotype but show no evidence of recurring recombination, allowing each distinct haplotype to be equated with an allele, all copies of which are essentially identical by descent. Historic recombinants, representing rare events that have drifted to common frequencies over many generations, can be identified in some cases, they do not equate to frequently recurring recombination. We have identified examples in our data collected on various projects and present eight such mini-haplotypes comprised of informative SNPs. We also discuss the ideal characteristics and advantages of minihaps for human familial identification and ancestry inference, and compare them to other types of forensic markers in use and/or that have been proposed. We expect that it is possible to carry out a systematic search and identify a useful panel of mini-haplotypes, with even better properties than the examples presented here.
Keywords: individual identification, SNP, haplotypes, minihaps, populations, ancestry inference
Introduction
Small molecular regions (for example, spanning <10 KB) comprised of three or more SNPs that define multi-allelic haplotype loci (minihaps) have the potential to convey more identity and ancestry-related information than a like number of single SNPs would convey. We defined and have advocated developing such multi-SNP haplotype systems as one type of forensic DNA marker, lineage informative SNPs, LISNPs (Pakstis et al,1 Butler et al;2 see also Ge et al3). The multiple alleles (haplotypes) available in these more complex systems can serve to identify relatives with higher probabilities than simple di-allelic SNPs. By restricting the molecular extent to under ∼10 KB in regions with no recombination hot spot, recombination among the SNPs will be so rare that the possibility of recombination within a kindred approaches the mutation rate for SNPs. Depending on allele frequency variation among populations, minihaps could also be useful in ancestry inference. Direct comparison testing could also benefit from minihaps, but match probabilities will in general be population specific.
Ge et al3 described a strategy for identifying sets of SNPs with nearly complete to complete linkage disequilibrium (LD) among them. Although we agree that the objective of identifying haplotypes for forensic purposes is valid, we think their primary criterion of complete LD results in loci with lower than optimal heterozygosity. Based on our interest in haplotypes and their global patterns, we have recently begun pursuing the same objective of finding forensically useful haplotype systems for familial identification as they can have high heterozygosity, the relevant issue for familial identification. Our strategy is very different from that of Ge et al.3 We focus on finding very closely spaced SNPs in a region that is not a ‘hot spot' for recombination but also does not show strong linkage disequilibrium (LD). Our criteria are to find regions in which we can (1) clearly see multiple haplotypes that show conservation of evolutionary lineages that have evolved from the ancestral human haplotype and (2) show no evidence of recurring recombination (though there may be some historic recombinants that have drifted to sufficiently high frequency to become common haplotypes). Such regions are not necessarily ‘LD blocks' or ‘haploblocks', as those terms are usually used3, 4 and are not necessarily in strong LD. This approach of haplotype lineage identification is justified by and illustrated in our recent work on other projects in which we have identified the gene segments with multiple haplotypes (alleles) but that show no evidence of historic recombination or at most a single historic recombinant: ADH7;5 POLB;6 SLITRK1;7 CYP2E1;8 CYP2C8;9 ADH1B;10 OCA211 and TAS2R16.12
For minihaps to be useful in forensics, haplotype frequencies must be known and ideally there must be at least moderate heterozygosity in most populations of relevance in forensics. Given the highly diverse ancestries of the US population, and increasingly of European populations, our collection of population samples of diverse global origins (Table 1) is particularly appropriate for identifying forensically useful minihaps. We have applied our empirical screening to identify potentially useful mini-haplotypes in a subset of data that we have accumulated in past and ongoing research projects. In these genomic regions, we have accumulated dense SNP data; such dense SNP data are not available for most other global studies. This report presents eight minihaps that have emerged as examples. These exclude the examples referred to in the papers cited above.
Table 1. The 45 population samples studied (descriptions of the populations and samples can be found in ALFRED).
| World region | Population sample | N | World region | Population sample | N |
|---|---|---|---|---|---|
| Africa | Biaka | 70 | NW Siberia | Komi Zyrian | 47 |
| Mbuti | 39 | Khanty | 50 | ||
| Yoruba, Nigeria | 78 | SC Asia | Keralites, S India | 30 | |
| Ibo, Nigeria | 48 | NE Siberia | Yakut | 51 | |
| Hausa, Nigeria | 39 | Pacific Is. | Nasioi | 23 | |
| Chagga, Tanzania | 45 | Micronesians | 37 | ||
| Masai, Tanzania | 22 | East Asia | Laotians | 119 | |
| Sandawe, Tanzania | 40 | Cambodians | 25 | ||
| Zaramo, Tanzania | 39 | Chinese, SF area | 60 | ||
| Afro-Americans | 90 | Chinese, Taiwan | 49 | ||
| Ethiopians | 32 | Hakka, Taiwan | 41 | ||
| SW Asia | Yemenite Jews | 43 | Koreans | 54 | |
| Druze | 103 | Japanese | 51 | ||
| Samaritans | 41 | Ami, Taiwan | 40 | ||
| Ashkenazi | 83 | Atayal, Taiwan | 42 | ||
| Europe | Adygei | 54 | N America | Pima, Mexico | 53 |
| Chuvash | 42 | Maya, Yucatan | 52 | ||
| Hungarians | 92 | S America | Quechua, Peru | 22 | |
| Russians, Archangelsk | 34 | Ticuna | 65 | ||
| Russians, Vologda | 47 | Rondonian Surui | 47 | ||
| Finns | 36 | Karitiana | 57 | ||
| Danes | 51 | ||||
| Irish | 118 | ||||
| Euro–Americans | 92 |
Methods
We have studied routinely 45 population samples that were available for this screen. Table 1 displays the sample sizes (averaging 53 individuals) for each of the 45 populations (organized by geographical region of the world). For these populations, we have sufficient typing data to estimate haplotype frequencies with generally acceptable standard errors, at least for the common haplotypes. We restricted our search to SNPs that we have typed for all individuals in all 45 populations and that meet Hardy–Weinberg expectations in all populations. Our typing methods are described in detail elsewhere.5, 6, 7, 8, 9, 10, 11 As noted in those studies, an occasional typing failure persists after two attempts to obtain an acceptable result. Experience has shown that the yield from additional attempts at typing is extremely low. Those persistent failures are randomly distributed among individuals and populations and appear idiosyncratic chance or individual- and SNP-specific aspects of the DNA unrelated at the population level to the markers involved. Overall, the missing data rate (‘typing failure rate') for the SNPs in the eight minihaps presented here was 1.83%.
We identified mini-haplotypes with the following properties: clusters of three to five SNPs with overall molecular extents below 10 000 bp and median heterozygosities ≥0.5 for the 45 population samples studied. We chose 10 KB to provide an upper limit of about 10−4 for the recombination rate assuming a rough genome average of 1% per megabase and the absence of a recombination hot spot. We sought SNP clusters in which the average pairwise LD values across the populations were weak to moderate (that is, r2 ≤0.60) to help maximize the informativeness (heterozygosity) of the haplotypes.
Haplotype frequencies were estimated using the HAPLO program13 that implements the EM algorithm and calculates jackknife standard errors as well as binomial standard errors. Individual multi-SNP phenotypes can be unambiguously resolved into the haplotype-based genotype by observation whenever none or only one of the SNPs is heterozygous; only phenotypes heterozygous for more than one SNP have an ambiguous mapping to genotype. Larger percentages of unambiguous genotypes make the maximum likelihood estimates of the haplotype frequencies more accurate. The haplotype frequencies were used to estimate the linkage disequilibrium (LD) between any two pairs of alleles. Linkage disequilibrium was calculated using the commonly used r2 (Δ2 in Devlin et al14). Such descriptive statistics as the expected heterozygosities and linkage disequilibrium characteristics of the haplotypes were calculated for each minihap system for all the populations studied.
Results and Discussion
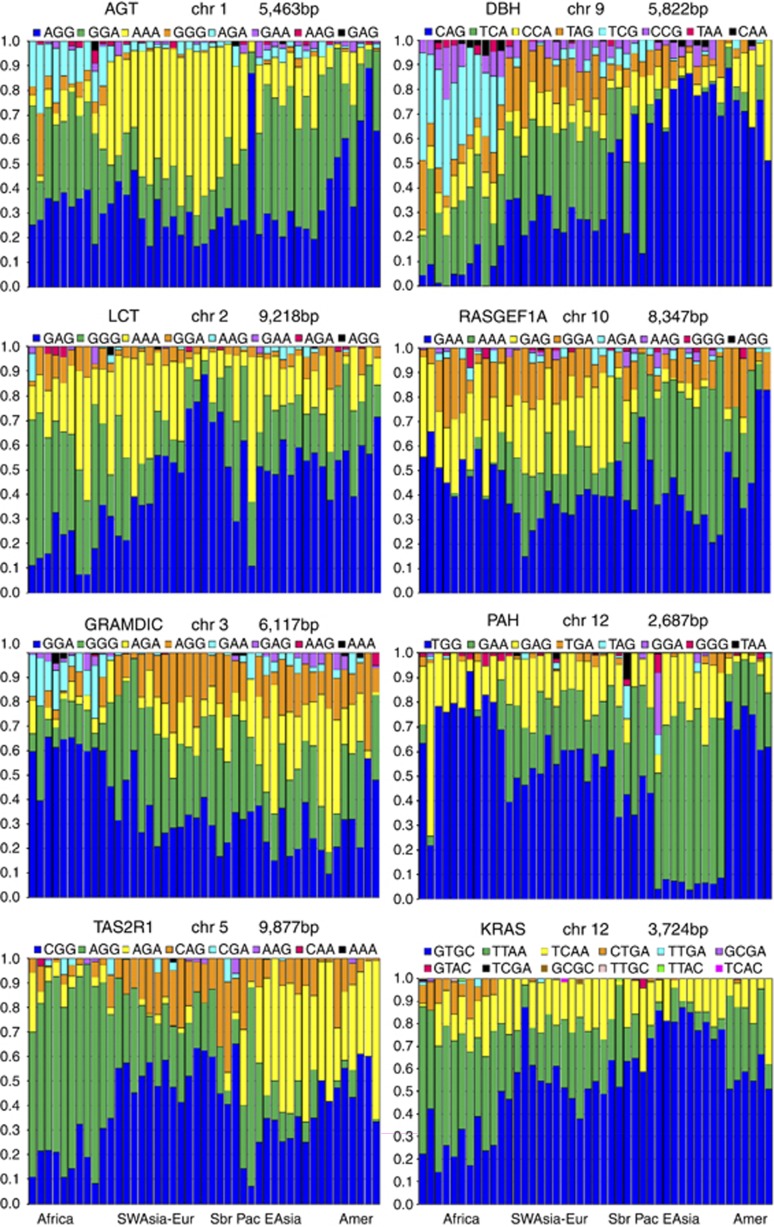
We examined over a dozen gene regions with dense SNP coverage and identified eight mini-haplotype systems that meet our screening criteria, seven regions defined by 3-SNPs and one defined by 4-SNPs. Table 2 presents the basic characteristics for these eight minihaps, including the molecular extent. For the eight minihaps we identified, Figure 1 presents the frequencies (in stacked bar format) of the individual haplotypes in each population. Allele frequencies have been deposited in ALFRED (with keyword ‘minihap') for each of the SNPs for the populations studied and for the minihaps; detailed results for each mini-haplotype in each population are also given in Supplemental Materials.
Table 2a. Characteristics of mini-haplotypes.
| Haplotype extent | LISNPs in chromosome order | Descriptive statistics: expected haplotype heterozygosity for N pops | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene region | Chr | (base pairs) | Leftmost SNP is toward pter | N | Avg | Median | Min | Max | ||
| AGT | 1 | 5463 | rs3789669 | rs699 | rs1078499 | 45 | 0.63 | 0.64 | 0.20 | 0.78 |
| LCT | 2 | 9218 | rs3213892 | rs1807356 | rs2304370 | 45 | 0.60 | 0.62 | 0.21 | 0.73 |
| GRAMD1C | 3 | 6117 | rs4422272 | rs7612534 | rs9865782 | 45 | 0.67 | 0.69 | 0.50 | 0.78 |
| TAS2R1 | 5 | 9877 | rs41462 | rs2234233 | rs41469 | 45 | 0.57 | 0.58 | 0.33 | 0.73 |
| DBH | 9 | 5822 | rs2519154 | rs739398 | rs77905 | 45 | 0.61 | 0.72 | 0.21 | 0.82 |
| RASGEF1A | 10 | 8347 | rs10899786 | rs4987092 | rs4987093 | 45 | 0.63 | 0.66 | 0.29 | 0.77 |
| PAH | 12 | 2687 | rs869916 | rs1722383 | rs1042503 | 45 | 0.51 | 0.54 | 0.14 | 0.74 |
| KRAS | 12 | 3724 | rs12587 | rs7973450 | rs9266, rs712 | 45 | 0.53 | 0.57 | 0.23 | 0.70 |
Figure 1.
Haplotype frequencies (proportional to colored bar lengths) in 45 populations from around the world. Each mini-haplotype consists of three LISNPs except for KRAS which has four LISNPs. Populations are ordered geographically on the x axis as in Table 1.
As can be seen from the minihap characteristics shown in Tables 2 and 3 and the allele frequencies for each of the eight minihaps across the populations studied, the screening was very successful. Each 3-SNP haplotype system has four to seven multi-SNP alleles (out of the eight possible haplotypes given three SNPs) with commonly occurring frequencies. The 4-SNP minihap has 2 to 4 haplotypes with common frequencies of 5% or more out of the 16 possible haplotypes. The median heterozygosity among the 45 populations studied ranges from 0.54 (PAH) to 0.72 (DBH) for the eight minihaps. Table 2b shows the percentage of the populations at each of the eight minihaps, with expected heterozygosities better than an optimally informative SNP (that is, ≥0.5). Although only one of the eight mini-haplotypes studied came close to having all population samples, with an expected heterozygosity better than an optimally heterozygous SNP (the GRAMD1C minihap has 44 of 45 populations with heterozygosity >0.5), five of the eight minihaps had over 87% (39 or more) of the population samples with heterozygosities exceeding that of an optimal SNP. For familial identification, heterozygosity is the most important variable as less common alleles are more informative in identifying a likely relative. For a diallelic SNP, the average allele frequency is 0.5; but for a multiallelic locus, the average allele frequency is 1/n. Although the informativeness of these minihaps tends to be good across all world regions, it would be desirable to have more stringent criteria that provide for a high minimum average heterozygosity for each world region. However, some of the smaller and especially the relatively isolated and inbred populations, such as the Karitiana and Nasioi, will naturally have lower average heterozygosities and require more effort if it is important to find good minihaps in those populations.
Table 2b. Characteristics of mini-haplotypes.
| Ancestral | Percentage of populations with mini-haplotype | Descriptive statistics: percentage of individuals in N pops with 0 or 1 heterozygotes for multi-SNP haplotypes (unambiguous phase) | |||||
|---|---|---|---|---|---|---|---|
| Gene region | haplotype | heterozygosity ≥0.5 (%) | N Pops | Avg% | Median% | Min% | Max% |
| AGT | AGA | 91 | 45 | 50 | 46 | 25 | 83 |
| LCT | GAG | 87 | 45 | 61 | 62 | 26 | 89 |
| GRAMD1C | GAA | 98 | 45 | 77 | 79 | 47 | 98 |
| TAS2R1 | AGG | 80 | 45 | 73 | 77 | 34 | 98 |
| DBH | TAG | 69 | 45 | 61 | 58 | 27 | 91 |
| RASGEF1A | GAG | 89 | 45 | 80 | 80 | 56 | 100 |
| PAH | TGG | 57 | 45 | 61 | 59 | 10 | 90 |
| KRAS | TTAA | 73 | 45 | 54 | 52 | 36 | 80 |
Table 2b also shows that a high proportion of the individuals with these 3-site and 4-site phenotypes in our eight mini-haplotype examples can be resolved unambiguously into haplotypic genotypes by direct examination because no more than one SNP is heterozygous. The median percentage of individual 3-SNP phenotypes is resolvable by direct examination in the populations studied (that is, unambiguous, ranges from 50 to 80%). This high percentage of resolvable genotypes will usually allow high probabilities for statistical estimation of the genotypes of the individuals with ambiguous phenotypes.
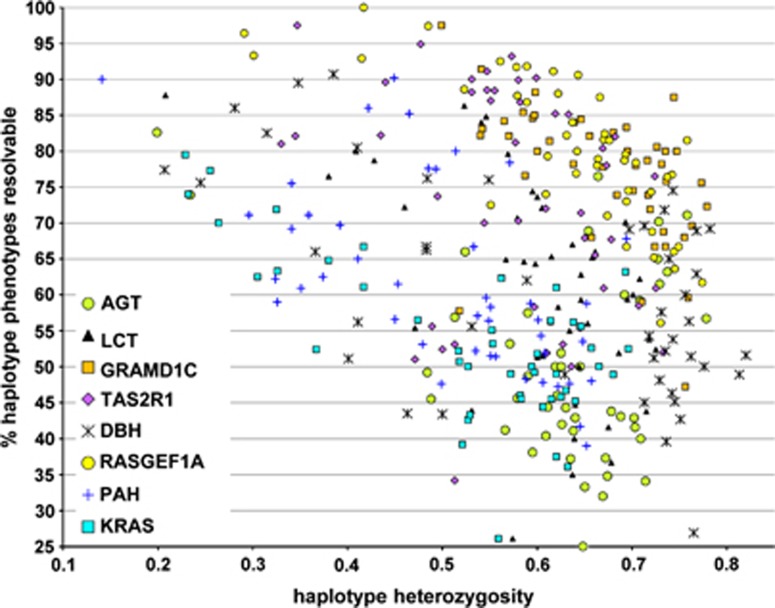
Each of the minihaps has different properties in each of the 45 populations. Figure 2 plots these 360 different situations by the heterozygosity of the system and the percent of individuals with unambiguous resolution of genotype. Both higher heterozygosity and a higher percentage of unambiguous resolution of genotype are better for familial inference. It is clear that the different loci differ in general and with respect to specific populations. Some minihaps, such as those at GRAMD1C, TAS2R1 and RASGEF1A, have both high heterozygosity (>0.5) and high resolvability (>0.5) in most populations (upper right quadrant). Others, especially at AGT, have high heterozygosity but low resolvability (<0.5) in most populations. Still others, such as at KRAS, show considerable variation among populations. Thus, it is obvious that the value of any minihap for familial inference is a function of the population in which it is studied. At the same time, the minihap data themselves can often help identify the relevant population to the degree that allele frequencies vary among populations. The informativeness of a locus for ancestry inference is a function of the allele frequency variation among populations, frequently measured as Fst. In the Supplementary Table S1, we present the Fst values for these eight minihaps and for the component individual SNPs. These loci were selected to be multiallelic in most populations biasing against high levels of allele frequency variation and show a range of Fst values distributed across the range we have seen for a random selection of individual SNPs in these populations.
Figure 2.
A scatterplot of each of the eight minihaps in each of the 45 populations plotting the heterozygosity and the percentage of individuals with an unambiguous genotype. The eight different loci are indicated with different symbols, as indicated. One PAH point (73% heterozygosity and 10% resolvable) falls outside the range visualized.
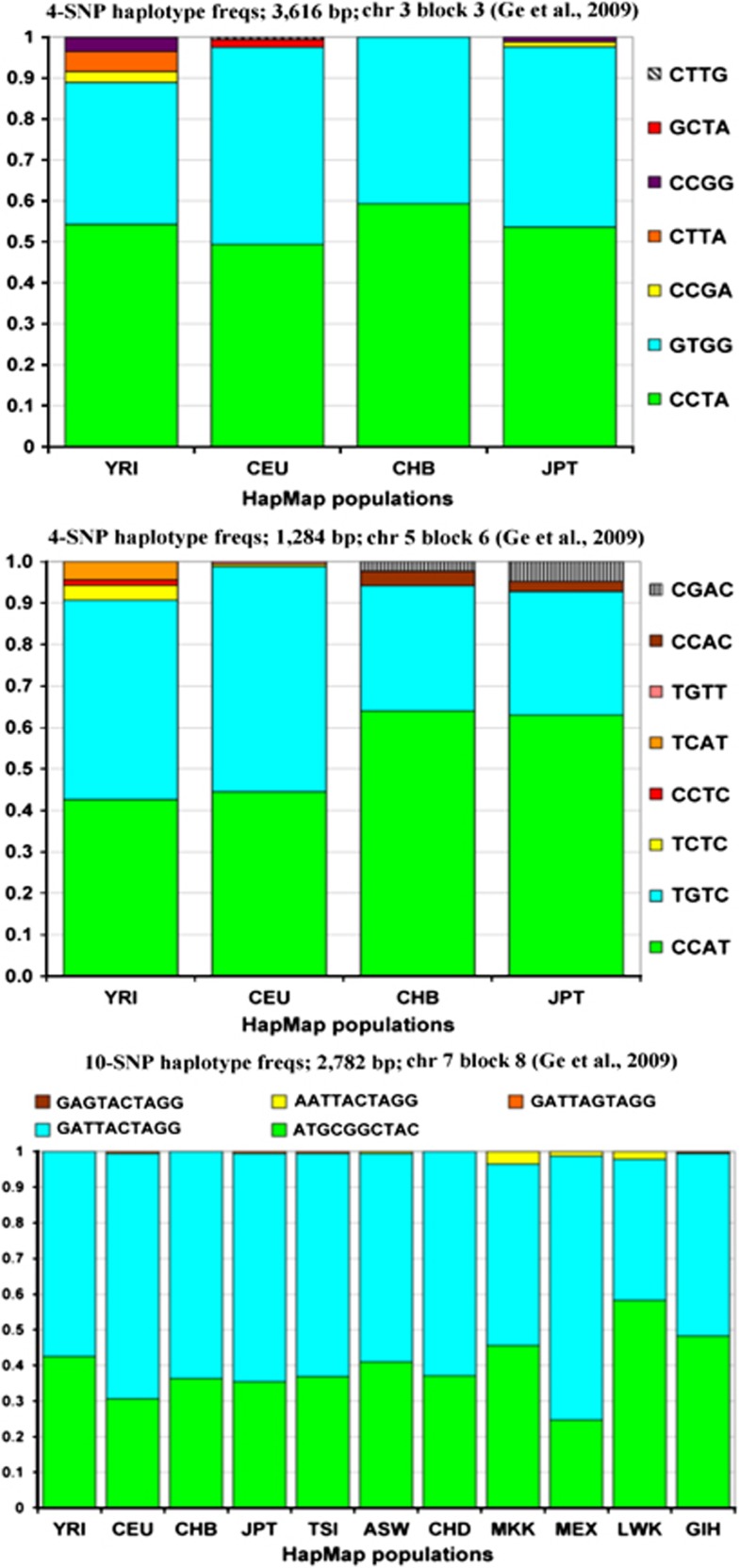
For comparison, we have also carried out analyses on the 24 ‘haploblocks' identified by Ge et al3 in the HapMap data set; the ‘haploblock' frequency bar graphs are shown in Figure 3 for three typical blocks (the details for the other 21 ‘haploblocks' can be found in the Supplemental Material). Based on this result, we conclude that the definition and the search procedure they described were not optimal, and identified many haplotypes that are only slightly better than a single SNP with maximum allele frequencies of 0.5 (Table 3, Figure 3, and Supplemental Data). This falls short of the potential informativeness for familial identification of molecularly short multi-SNP haplotype systems and mitigates the advantage of using haplotypes. Our definition and search procedures, as shown by our minihap examples, result in higher heterozygosity on average than is possible with an individual SNP, demonstrating the validity of the minihap concept and its distinction from the ‘haploblock' concept. Although heterozygosity is a function of actual haplotype frequencies, a hypothetical 3-SNP haplotype system with five of eight possible haplotypes at equal frequencies will have a heterozygosity of 0.80 (if all eight possible haplotypes occur at equal frequencies, the heterozygosity is 0.875).
Figure 3.
Frequency bar graphs for 3 of the 24 ‘haploblock' haplotypes identified by Ge et al3 using HapMap information. Compare to bar graphs in Figure 1 for the eight minihaps.
Table 3. Multi-SNP haplotypes: heterozygosity and pairwise LD values (r 2) for all SNP pairs.
| ‘Haploblock' | #SNPs | #Pops | # LD | Min | Max | Avg | Avg Hap. | % Pops | # Haps with Frqs ≥5% | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Geet al, 2010 | studied | Hap Map | values | LD | LD | LD | Het. | ≥ 0.5 Het | Min | Med | Max |
| Chr 2 block 1 | 4 | 11 | 66 | 0.63 | 1.00 | 0.91 | 0.53 | 100 | 2 | 2 | 3 |
| Chr 3 block 2 | 3 | 11 | 33 | 0.47 | 1.00 | 0.84 | 0.48 | 36 | 2 | 3 | 3 |
| Chr 3 block 3 | 4 | 4 | 24 | 0.59 | 1.00 | 0.91 | 0.53 | 75 | 2 | 2 | 3 |
| Chr 4 block 4 | 3 | 11 | 33 | 0.70 | 1.00 | 0.95 | 0.50 | 46 | 2 | 2 | 3 |
| Chr 5 block 5 a | 6 | 11 | 165 | 0.55 | 1.00 | 0.88 | 0.54 | 82 | 2 | 2 | 3 |
| Chr 5 block 5 b | 12 | 4 | 264 | 0.78 | 1.00 | 0.96 | 0.53 | 75 | 2 | 2 | 2 |
| Chr 5 block 6 | 4 | 4 | 24 | 0.72 | 1.00 | 0.87 | 0.53 | 75 | 2 | 2 | 2 |
| Chr 5 block 7 | 2 | 11 | 11 | 0.43 | 0.95 | 0.67 | 0.56 | 91 | 2 | 3 | 4 |
| Chr 7 block 8 | 10 | 11 | 495 | 0.87 | 1.00 | 0.99 | 0.47 | 27 | 2 | 2 | 2 |
| Chr 7 block 9 | 3 | 4 | 12 | 0.58 | 1.00 | 0.90 | 0.50 | 50 | 2 | 2 | 3 |
| Chr 8 block 10 | 3 | 4 | 12 | 0.77 | 1.00 | 0.88 | 0.54 | 100 | 2 | 2 | 2 |
| Chr 8 block 11 | 3 | 11 | 33 | 0.53 | 1.00 | 0.84 | 0.51 | 55 | 2 | 2 | 3 |
| Chr 10 block 12 | 4 | 4 | 24 | 0.77 | 1.00 | 0.88 | 0.48 | 25 | 2 | 2 | 3 |
| Chr 11 block 13 | 6 | 4 | 60 | 0.70 | 1.00 | 0.91 | 0.42 | 0 | 2 | 2 | 2 |
| Chr 11 block 14 | 3 | 4 | 12 | 0.70 | 1.00 | 0.86 | 0.50 | 75 | 2 | 2.5 | 3 |
| Chr 11 block 15 | 3 | 4 | 24 | 0.58 | 1.00 | 0.88 | 0.53 | 75 | 2 | 2.5 | 3 |
| Chr 12 block 16 a | 2 | 4 | 4 | 0.82 | 1.00 | 0.92 | 0.50 | 50 | 2 | 2 | 2 |
| Chr 12 block 16 b | 3 | 11 | 33 | 0.68 | 1.00 | 0.90 | 0.51 | 73 | 2 | 2 | 3 |
| Chr 13 block 17 | 2 | 4 | 24 | 0.82 | 1.00 | 0.96 | 0.51 | 75 | 2 | 2 | 2 |
| Chr 13 block 18 | 3 | 11 | 33 | 0.67 | 1.00 | 0.87 | 0.52 | 73 | 2 | 3 | 3 |
| Chr 13 block 19 a | 3 | 4 | 12 | 0.94 | 1.00 | 0.99 | 0.43 | 0 | 2 | 2 | 2 |
| Chr 13 block 19 b | 3 | 8 | 24 | 0.61 | 1.00 | 0.81 | 0.50 | 38 | 2 | 3 | 3 |
| Chr 14 block 20 | 3 | 4 | 12 | 0.69 | 1.00 | 0.84 | 0.55 | 75 | 2 | 2.5 | 3 |
| Chr 14 block 21 | 3 | 4 | 12 | 0.67 | 1.00 | 0.84 | 0.55 | 100 | 2 | 3 | 3 |
| Chr 14 block 22 | 5 | 4 | 40 | 0.73 | 1.00 | 0.86 | 0.54 | 50 | 2 | 3 | 3 |
| Chr 16 block 23 | 3 | 4 | 12 | 0.72 | 1.00 | 0.86 | 0.51 | 50 | 2 | 2.5 | 3 |
| Chr 18 block 24 | 4 | 4 | 24 | 1.00 | 1.00 | 1.00 | 0.47 | 25 | 2 | 2 | 2 |
| Mini-haplotypes | #LD | Min | Max | Avg | Avg Hap. | %Pops | # Haps with Frqs ≥5% | ||||
|
in this paper |
#SNPs |
#Pops |
values |
LD |
LD |
LD |
Het. |
≥0.5 Het. |
Min |
Med |
Max |
| AGT | 3 | 45 | 135 | 0.00 | 1.00 | 0.39 | 0.63 | 91 | 2 | 3 | 5 |
| LCT | 3 | 45 | 135 | 0.00 | 1.00 | 0.32 | 0.60 | 87 | 2 | 3 | 4 |
| GRAMD1C | 3 | 45 | 113 | 0.00 | 0.87 | 0.05 | 0.67 | 98 | 2 | 4 | 5 |
| TAS2R1 | 3 | 45 | 127 | 0.00 | 1.00 | 0.17 | 0.57 | 80 | 2 | 3 | 5 |
| DBH | 3 | 45 | 131 | 0.00 | 1.00 | 0.26 | 0.61 | 69 | 1 | 4 | 6 |
| RASGEF1A | 3 | 45 | 121 | 0.00 | 0.30 | 0.06 | 0.63 | 89 | 2 | 4 | 5 |
| PAH | 3 | 45 | 123 | 0.00 | 1.00 | 0.55 | 0.51 | 57 | 2 | 3 | 5 |
| KRAS | 4 | 45 | 270 | 0.00 | 1.00 | 0.63 | 0.53 | 73 | 2 | 3 | 4 |
Notes: some LD values could not be computed because in some population samples one or both of the paired alleles does not vary. The 11 Phase 3 HapMap populations include: the original four populations (YRI, Yoruba from Ibadan Nigeria; CEU, the CEPH panel of descendants of northwestern Europeans in Utah; CHB, Chinese from Beijing; and JPT, Japanese from Tokyo) plus seven additional population samples (TSI, Tuscans from Italy; ASW, individuals of African ancestry from the southwest US; CHD, Chinese from the Denver area; MKK, Maasai from Kenya; MEX, a sample of Mexican ancestry from Los Angeles; LWK, Luhya from Kenya; and GIH, Indians from Gujarat sampled in Houston, Texas).
For use in familial searching or ancestry inference, it is important that the ‘mutation rate' be very low. In the case of haplotypes of SNPs, that translates into a very low recombination rate since we already know mutation rates for individual SNPs are on the order of 10−8 (Reich et al15). This is significantly lower than the estimated mutation rates at STRPs.16, 17 Thus, we expect that a new mutation within a family at one of the relevant SNPs (or affecting a PCR primer function) will be very low, even considering the multiple meioses that may be involved. Recurring recombination in the minihaps we have identified is very rare to be absent in human populations globally. The selection criteria and the data support the validity of assuming that the haplotypes identified and being studied are identical by descent over most of recent human evolution. Obviously, it is necessary to avoid regions that encompass a recombination hot spot as we have identified at other loci.5, 8 However, high frequency of a recombinant haplotype does not necessarily indicate a hot spot for recombination. A single historic crossover gamete may have drifted to high frequency over many human generations. Thus, the actual haplotype frequencies depend on the pattern of accumulated mutations and any historical crossovers plus the historical random genetic drift making the new haplotypes sufficiently common.
It is important to recognize that measures of LD are not measures of recombination rate but of recombinant haplotypes in the population. Thus, the presence of all four combinations of the alleles at two di-allelic SNPs gives an LD value less than 1.0, but that does not mean there is recurring recombination. One check for the relative frequency of recombination is to examine the flanking SNPs. Occurrence of many different combinations among the SNPs on either side of an interval, as is seen at ADH7,5 is an indication of frequent recombination. In contrast, very similar flanking combinations (allowing for mutation) relative to the targeted genomic interval, as is seen at ADH1B,10 likely indicate a single origin from a historic crossover. For each pairwise comparison there should be at least three of the four possible allelic combinations present in order for r2 to yield a value less than 1.0 (though D' will be exactly 1.0 if only three combinations occur). If both r2 and D' equal 1.0 then only two combinations occur and one of the two SNPs is completely redundant (that is, uninformative). This is apparently the case for the ‘haploblocks' identified by Ge et al.3
As seen in Figure 1, and our studies of various genes cited earlier, some requisite recombination events appear to have occurred and the resulting haplotype became common early in human dispersal. These recombinant haplotypes may have persisted as identical by descent (IBD) lineages since their origin. Thus, these historical recombination events can generate haplotypic heterozygosity but do not necessarily indicate high recombination rates, per se, just high frequency of one or a few crossover products. We note that assumption of equilibrium and use of frequency of recombinant chromosomes to estimate recombination rates can give different rates for different populations, none of which may be correct. The distinction needs to be considered in any search for forensically useful minihaps.
We think that this report substantiates the feasibility of finding a panel of multi-allelic mini-haplotypes that would be useful in routine forensic applications in many different populations from around the world. Substantial SNP resources exist (for example, the Human Genome Diversity Project 650Y Illumina chip data set among others) to search for more and better mini-haplotypes. We note the caveat that SNPs in the HGDP data set are often not sufficiently dense for minihap forensic purposes, although there is an inverse relationship with actual recombination rate—in a region of very low recombination the markers can be more distant. SNP data in the HapMap data sets are much more dense, but the representation of global human diversity is much poorer. The increasing amount of genomic sequence data provides another resource for identification of minihaps, but as yet global human diversity is poorly represented.
The population data we provide in this paper are essential for any use of minihaps in forensic applications: calculating the probability of a match between an individual and crime-scene DNA, inference of the likely ancestry of the source of a DNA sample and inference of the likelihood that a relative of a known person might match a similar crime-scene sample (that is, familial searching). This last case is also applicable in mass disaster and missing persons cases in which data on an individual (or on ‘remains') need to be assigned to families. All of these calculations will depend on estimating the population-specific haplotype-based genotype from the multisite phenotype when the phenotype is ambiguous. Nonetheless, we note that most phenotypes are not ambiguous (cf., Figure 2) and even some of the ambiguous phenotypes have very large probabilities of logically mapping to a single genotype. In all cases, but especially in the familial searching (LISNP) context, it is necessary to calculate likelihood ratios considering the probabilities of a match, of an ancestry, or of a family member identification relative to the probabilities of the phenotype occurring by chance in each specific relevant population. In the mass disaster case, the relative likelihoods of the assignment to different families would need to be calculated. All of these estimates are population dependent and can be estimated from the population haplotype frequencies presented here. Basic Mendelian transition probabilities18 can be used in combination with the phenotype to genotype probabilities to determine the numerator in the familial searching and mass disaster situations.
The eight examples we present here are statistically independent both in terms of LD within populations and in terms of linkage between minihaps, even the two on chromosome 12 (∼75 MB apart on separate arms), allowing simple multiplication among the values calculated for the individual minihaps. However, we are not advocating immediate attempts to implement this panel as we consider these eight to be insufficient in general for highly significant statistical results in any of the applications above. Nonetheless, a likelihood ratio could be quite large in a specific population for some specific combination of genotypes among relatives. These eight minihaps do illustrate the concept and provide the conceptual basis for actual application in forensics considered broadly to include identification of family relationships and ancestry. Ultimately, they can be used along with other minihaps to be identified and documented in future studies. These examples are also centered around known genes, though none of the sites is part of an expressed protein. For some researchers/ethicists this could raise an issue of whether privacy and personal health information are compromised. We argue that these SNPs are all normal variation as documented by their high heterozygosity and the multiple haplotypes common in all populations. Hence, none could be strongly associated with a highly deleterious trait. Thus, until and unless some exceedingly high risk of disease/disorder is demonstrated–a very unlikely event given the population genetics of these minihaps–we consider these to be ethically acceptable forensic markers.
In summary, we have shown how, by using certain selective criteria, mini-haplotypes can provide the informativness of multiallelic loci, while using SNP genotyping technology. Moreover, these minihaps have the evolutionary stability that allows haplotypes to be equated with alleles basically identical by descent in broader studies. Because of their high potential heterozygosity and the population differences in haplotype frequencies, we expect that mini-haplotypes are likely to be very useful markers for connecting an individual to an extended family or clan in forensic work. Thus, they fall into the lineage-informative category of genetic markers. They can also provide information on ancestry, as shown by the AAA haplotype at AGT (most frequent in Europeans) and by the GAA haplotype at PAH (most frequent in East Asians). Information from such markers may be especially valuable for identification purposes in the case of mass disasters.
Acknowledgments
This work was funded primarily by Grants 2007-DN-BX-K197 and 2010-DN-BX-K225 to KKK awarded by the National Institute of Justice, Office of Justice Programs, US Department of Justice. Points of view in this document are those of the authors and do not necessarily represent the official position or policies of the US Department of Justice. Much of the original data collection was supported by GM057672 from the US National Institutes of Health. We would also like to thank all the collaborators who helped to collect the samples as well as the National Laboratory for the Genetics of Israeli Populations at Tel-Aviv University and the Coriell Cell Repositories. Special thanks to hundreds of individuals who volunteered to give blood samples for studies of gene frequency variation.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Pakstis AJ, Speed WC, Kidd JR, Kidd KK.An expanded, nearly universal, panel of SNPs for individual identification Poster Presented at Annual NIJ Meeting 2007. available online: http://medicine.yale.edu/labs/kidd/www/NIJposter2007.pdf .
- Butler JM, Budowle B, Gill P, et al. Report on ISFG SNP panel discussion. Forensic Sci Int. 2008;1:471–472. [Google Scholar]
- Ge J, Budowle B, Planz JV, Chakraborty R. Haplotype block: a new type of forensic DNA markers. Int J Legal Med. 2010;124:353–361. doi: 10.1007/s00414-009-0400-5. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Han Y, Gu S, Oota H, et al. Evidence of positive selection on a class I ADH locus. Am J Hum Genetics. 2007;80:441–456. doi: 10.1086/512485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamtich J, Speed WC, Straka E, Kidd JR, Sweasy JB, Kidd KK. Population-specific variation in haplotype composition and heterozygosity at the POLB locus. DNA Repair. 2009;8:579–584. doi: 10.1016/j.dnarep.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed WC, O'Roak BJ, Tarnok Z, Barta C, Pakstis AJ, State MW, Kidd KK. Haplotype evolution of SLITRK1, a candidate gene for Gilles de la Tourette syndrome. Am J Med Genet B Neuropsychiatric Genet. 2008;147B:463–466. doi: 10.1002/ajmg.b.30641. [DOI] [PubMed] [Google Scholar]
- Lee MY, Mukherjee N, Pakstis AJ, et al. Global patterns of variation in allele and haplotype frequencies and linkage disequilibrium across the CYP2E1 gene. Pharmacogenomics J. 2008;8:349–356. doi: 10.1038/tpj.2008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed WC, Kang SP, Tuck DP, Harris LN, Kidd KK. Global variation in CYP2C8-CYP2C9 functional haplotypes. Pharmacogenomics J. 2009;9:283–290. doi: 10.1038/tpj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Gu S, Han Y, et al. Diversification of the ADH1B gene during expansion of modern humans. Ann Hum Genetics. 2011;75:497–507. doi: 10.1111/j.1469-1809.2011.00651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly MP, Paschou P, Grigorenko E, et al. The distribution and most recent common ancestor of the 17q21 inversion in humans. Am J Hum Genetics. 2010;86:161–171. doi: 10.1016/j.ajhg.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Pakstis AJ, Kidd JR, Kidd KK. Selection on the human bitter taste gene, TAS2R16, in Eurasian populations. Hum Biol. 2011;83:363–377. doi: 10.3378/027.083.0303. [DOI] [PubMed] [Google Scholar]
- Hawley M, Kidd KK. HAPLO: a program using the EM algorithm to estimate the frequencies of multi-site haplotypes. J Heredity. 1995;86:409–411. doi: 10.1093/oxfordjournals.jhered.a111613. [DOI] [PubMed] [Google Scholar]
- Devlin B, Risch N. A comparison of linkage disequilibrium measures for fine scale mapping. Genomics. 1995;29:311–322. doi: 10.1006/geno.1995.9003. [DOI] [PubMed] [Google Scholar]
- Reich DE, Schaffner SF, Daly MJ, et al. Human genome sequence variation and the influence of gene history, mutation and recombination. Nat Genet. 2002;32:135–140. doi: 10.1038/ng947. [DOI] [PubMed] [Google Scholar]
- Huang QY, Xu FH, Shen H, et al. Mutation patterns at dinucleotide microsatellite loci in humans. Am J Hum Genetics. 2002;70:625–634. doi: 10.1086/338997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy BM, Stenersen M, Egeland T, Olaisen B. Y-chromosomal microsatellite mutation rates: differences in mutation rate between and within loci. Hum Mutat. 2004;23:117–124. doi: 10.1002/humu.10294. [DOI] [PubMed] [Google Scholar]
- Li CC, Sacks L. The derivation of joint distribution and correlation between relatives by use of stochastic matrices. Biometrics. 1954;10:347–360. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.