Figure 2.
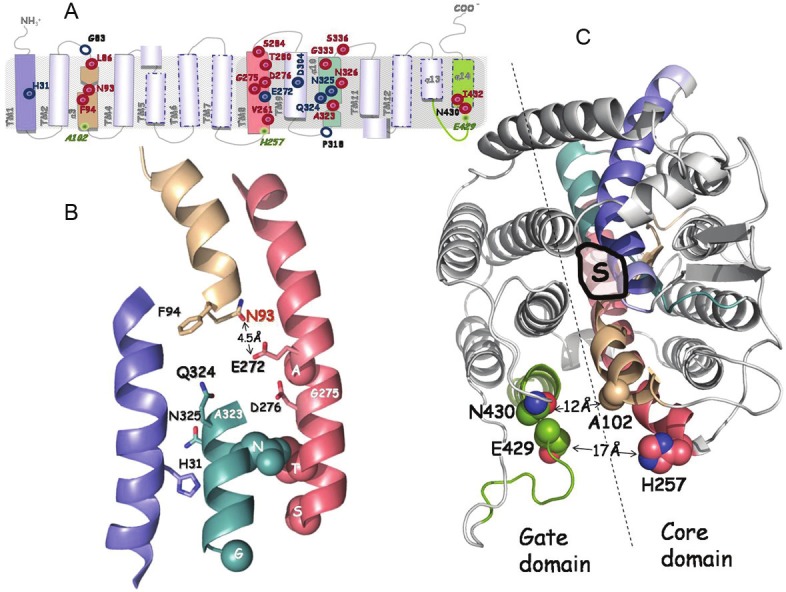
Synopsis of current knowledge on the structure-function analysis of xanthine permease XanQ. The positions of key residues of XanQ are shown in models depicting topology (A), arrangement of the four helices forming the substrate shelter (B) and the overall two-domain structure of the permease (C), as deduced from homology threading on the template of UraA [25,31]. Different colors indicate transmembrane segments TM1 (blue), TM3 (wheat), TM8 (salmon), TM10 (teal) and TM14 (split pea green). In panel A, important residues are indicated with empty blue circles (irreplaceable for expression in the membrane), blue targets (irreplaceable for uptake activity as well as the affinity-related H31) or red targets (sensitive to alkylation or replacement with several side chains). Positions studied for intramolecular interactions are indicated as miniscule green targets. In panel B, the side chains of important residues are shown as thick sticks (irreplaceable and/or substrate binding-relevant) or spheres (sensitive to alkylation or bulky replacement). Residues G275 and A323 where sensitivity to alkylation is enhanced by substrate are indicated with a white label. The specificity-related N93 is shown with red label. In panel C, shown are the coordination site of substrate (S) and distances between residues of the core and the gate domains at the periplasmic side. Further details can be found in the text.
