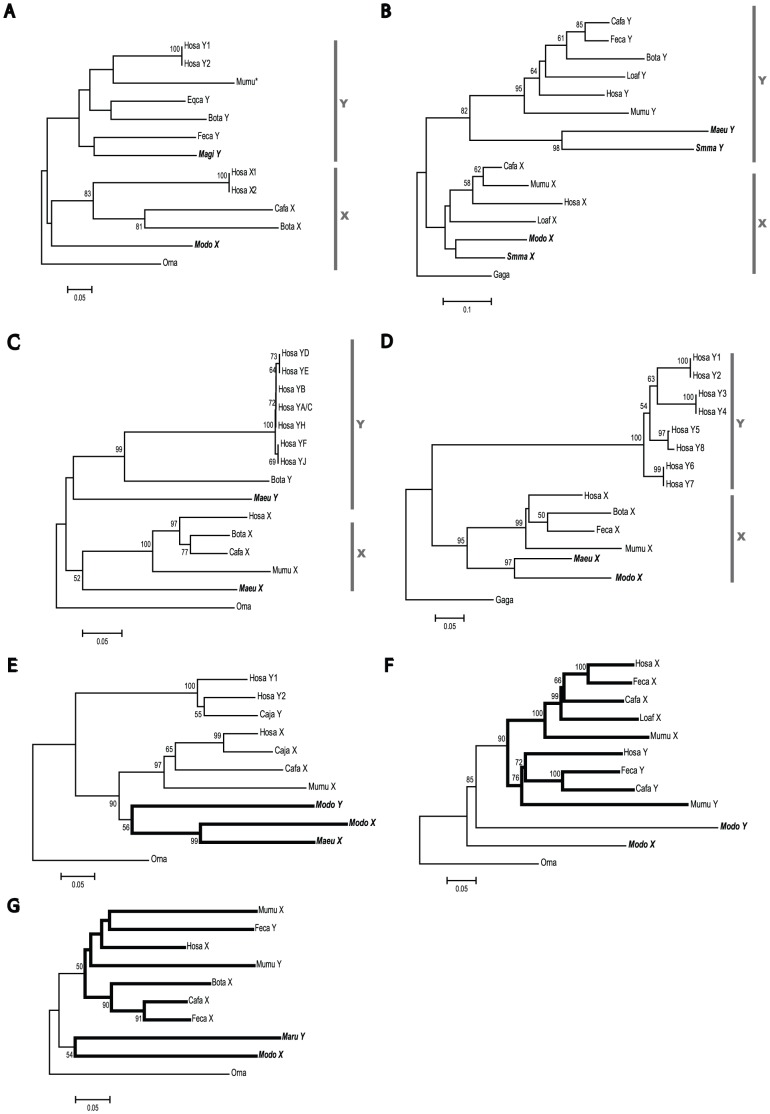
Figure 2. The phylogenetic relationships of 7 gametologs.
Neighbor-joining trees were constructed on the basis of the number of synonymous differences per site (p S). The bootstrap value supporting each internal branch is indicated at the node. Only a bootstrap value of more than 50% is shown. Sequences used for tree construction are listed in Table S1. The number of synonymous sites compared (excluding gaps) and that of operation taxonomy units (OTUs) are as follows: (A) HSFX/Y (96 sites; 13 OTUs), (B) SOX3/SRY (70 sites; 15 OTUs), (C) RBMX/Y (289 sites; 15 OTUs), (D) XKRX/Y (114 sites; 15 OTUs), (E) RPS4X/Y (289 sites; 11 OTUs), (F) SMCX/Y (1280 sites; 12 OTUs), and (G) UBE1X/Y (147 sites; 10 OTUs). Platypus sequences were used as an outgroup, except in trees B and D. For trees B and D, chicken sequences were used as an outgroup. A vertical gray bar beside each tree shows a monophyletic cluster of X- or Y-linked genes. Bold branches in E, F, and G show either marsupial- or eutherian-specific clusters. OTU names in bold indicate marsupials. The abbreviation for species names are as follows: Bota (Bos taraus), Cafa (Canis familiaris), Caja (Callithrix jacchus), Eqca (Equus caballus), Feca (Felis catus), Gaga (Gallus gallus), Hosa (Homo sapiens), Loaf (Loxodonta africana), Maeu (Macropus eugenii), Magi (Macropus giganteus), Maru (Macropus rufus), Modo (Monodelphis domestica), Mumu (Mus musculus), Orna (Ornithorhynchus anatinus), and Smma (Sminthopsis macroura). Mumu* in HSFX/Y tree (A) is located on chromosome 1 (see Discussion). BotaY sequence was not included in the UBE1X/Y tree (G) because it is truncated (Fig. S1).