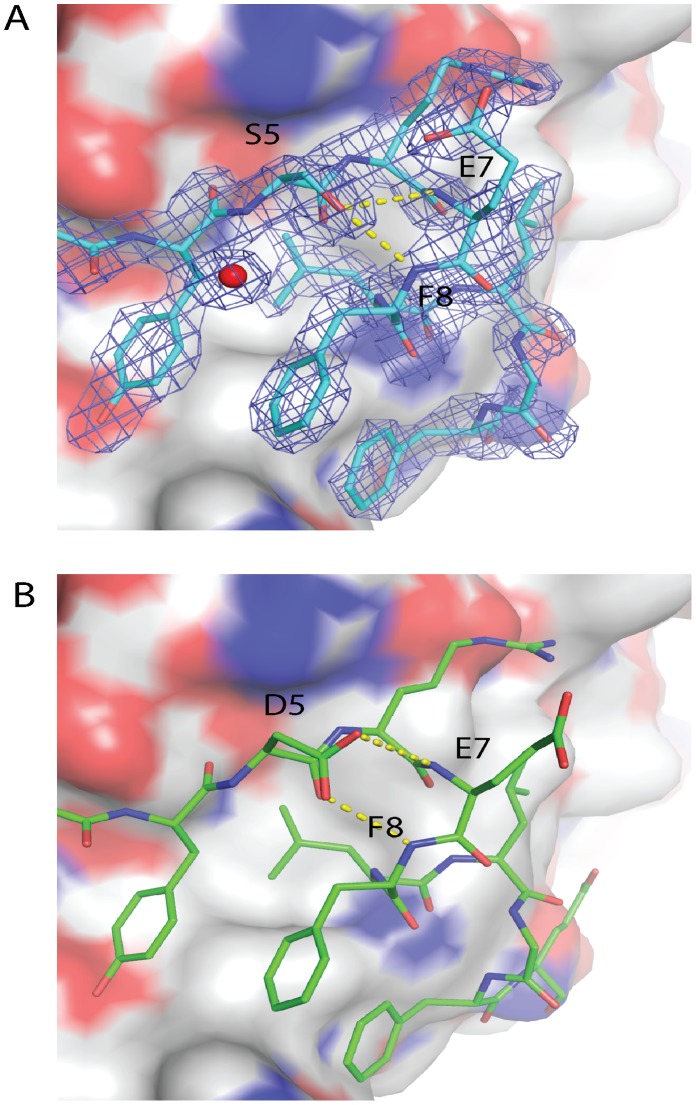
Figure 2. Structural comparison of N-Capping motifs either containing S or D at position 5 in eIF4E interacting peptides when bound to eIF4E. A).
Crystal structure of the eIF4G1-D5S derivative peptide bound to eIF4E. It can be seen that S5 makes two optimized hydrogen bonds to the amide backbone groups of R6 and E7 located in the first turn of the helix of the bound peptide. The electron density for the eIF4G1-D5S peptide in the 2Fo-Fc map is shown with the blue mesh and is contoured at 1.5σ. B) D5 in the 2W97 structure bound to the eIF4G1 wild type peptide also makes these two hydrogen bonds but their geometry is not as optimal as that seen for the hydrogen bonds formed in the eIF4G1_D5S peptide.