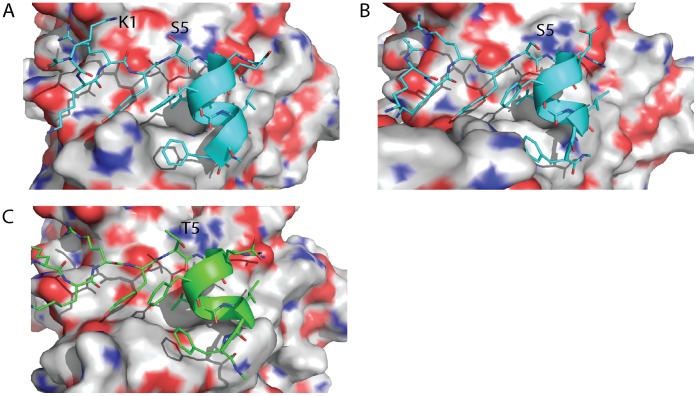
Figure 3. Comparison of the structural dynamics of N-Capping motifs, in peptides bound to eIF4E, containing either S or T at position 5.
The S5 and T5 N-Capping residues show distinctly different behaviors with respect to each other in terms of the frequency of rotation of their side chains in their respective simulations when bound to eIF4E. A) The S5 side chain can form an interaction with the K1 side chain of the bound peptide and point away from the helix leaving the amides solvated. B) The S5 side chain can also form hydrogen bond interactions with the free amide groups of the first turn of the peptide’s helix. C) The T5 side chain can also make these interactions, however if the hydroxyl of the T5 interacts with K1, its methyl group will disrupt solvation of the free amide groups. Thus it is energetically more favourable for T5 to align its hydroxyl group towards the helix whist S5, which lacks the methyl, has more freedom to rotate, and engages in one of the two hydrogen bonds. Deviations in the planarity of the tyrosine and phenylalanine ring systems are within the tolerances of the torsional restraints of the MD simulations. [31].