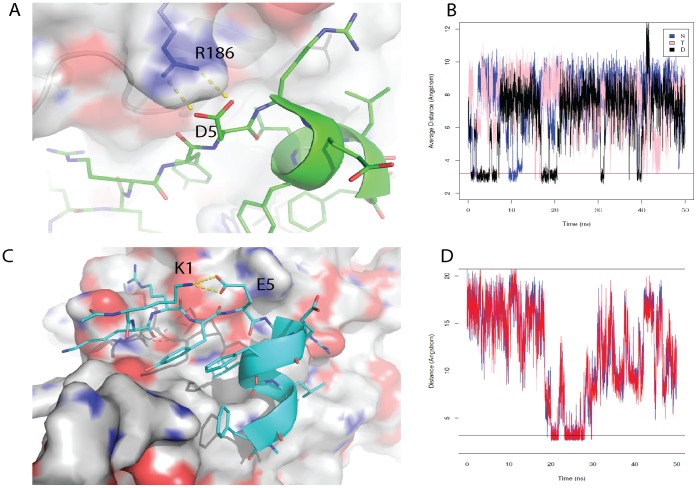
Figure 4. Detrimental interactions that attenuate peptide binding to eIF4E when D or E are incorporated at position 5. A).
Snapshot from the 50 ns computer simulation of the eIF4G1 wild-type peptide bound to eIF4E. In the eIF4G1 wild type peptide the D5 side chain is able to hydrogen bond with R186 of the receptor, deforming the N-terminal helix of the bound peptide. This deformation causes E7 of the peptide to interact more frequently with R6, which is also found on the peptide. This interaction disrupts the electrostatic interaction between R196 found on the surface of eIF4E and E7 of the eIF4G1 peptide, which helps to stabilize the formation of the peptide:protein complex. B) The average distance between E7 and R6 throughout the 50 ns simulations for the bound peptides eIF4G1-D5S, eIF4G1-T5S and the wild-type peptide. The average distance frequently dips below 3.2 Å for the wild type peptide compared to the S5 or T5 derivative peptides, which indicates that E7 and R6 interact more frequently with each other and destabilize complex formation in the wild type peptide compared to the derivative peptides. C) A snapshot from the 50 ns simulation of the complex between eIF4G1-D5E and eIF4E, showing the formation of a loop-like structure preceding the N-terminus of the peptide. The formation of the loop structure arises from the electrostatic interaction of E5 of the peptide with K2 at the N-terminus of the peptide. D) The distances between the two O atoms (the red and blue lines on the plot) of the E5 side chain and the N atom of the K1 side chain were plotted over the course of the simulation. The plot reveals that for a significant portion of the simulation these residues are within 3.2 Å of each other indicating the formation of a stable electrostatic interaction. This interaction hinders the interaction of K1 with E132 on the surface of eIF4E and leads to further destabilization of the eIF4E-peptide complex. Deviations in the planarity of the tyrosine and phenylalanine ring systems are within the tolerances of the torsional restraints of the MD simulations. [31].