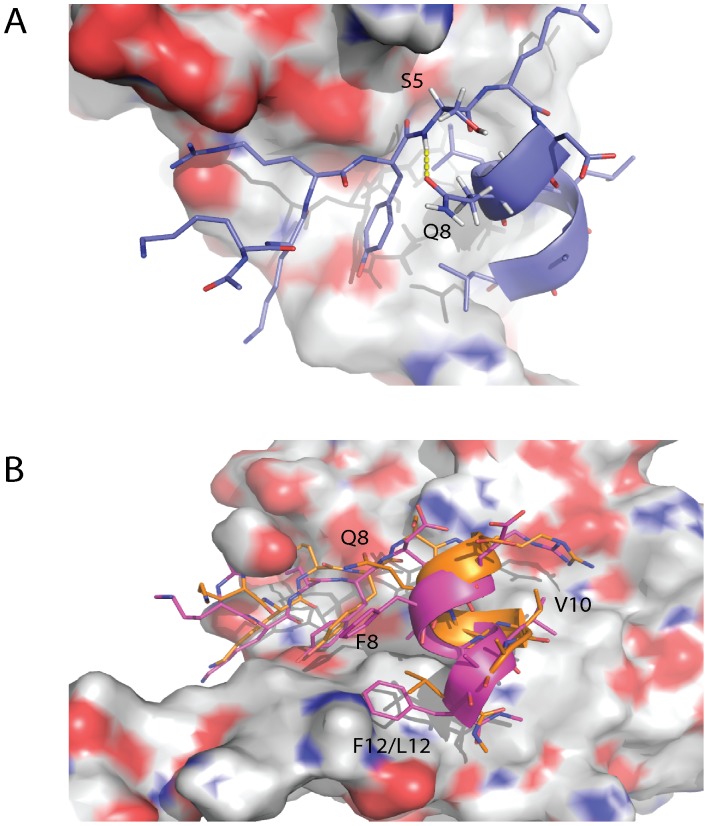
Figure 7. Q8 stabilizes the N-terminal end of the α-helix in eIF4E interacting peptides and facilitates the librational movement of the PHAGESOL peptide across the surface of eIF4E.
A) A representative snapshot showing the formation of an intra-molecular hydrogen bond between the side chain of Q8 and the backbone amide of S5. The formation of this hydrogen bond stabilizes the N-terminal portion of the helix in the bound peptide. The snapshot was taken from the simulation for the KKRYSRDQLLAL peptide bound to eIF4E. B) Overlay of representative snapshots from the PHAGESOL (KKRYSRDQLVAL) and eIF4G1-G11A (KKRYDREFLLAF) peptide simulations when bound to eIF4E. V10 of the PHAGESOL (orange) peptide orients itself and the helical turn it is located on, into a position where it can occupy the volume of space that would otherwise be occupied by the conserved L as shown here on the eIF4G1 derivative peptide (magenta). The movement of V10 across the surface of eIF4E results in the helical segment of the peptide spatially re-orientating itself in contrast to the other derivative peptides. This conformational change is facilitated by the presence of Q8 that stabilizes the N-terminal helix of the peptide and forms few interactions with eIF4E in contrast to F8 in eIF4G1-G11A. Deviations in the planarity of the tyrosine and phenylalanine ring systems are within the tolerances of the torsional restraints of the MD simulations. [31].