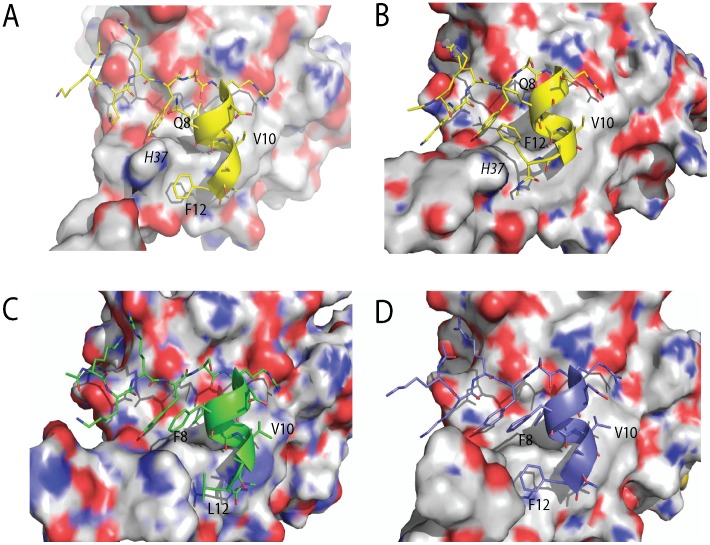
Figure 8. The presence of F8 impedes the structural fluctuations observed at the C-terminal of eIF4E interacting peptides.
When L12 is replaced by F12 to generate the PHAGESOL-L12F peptide, two contrasting interactions are formed with eIF4E. A) In the first interaction H37 of eIF4E can intercalate itself between Q8 and F12 of the peptide in the volume of space usually occupied by F8 in other derivative peptides. B) The second conformation sees the unwinding of the helical turn at the C-terminus of the peptide allowing F12 to partially occupy the space next to Q8 previously occupied by H37. H37 instead now forms a stacking interaction with F12. It is the result of these alternative packing arrangements that prevents V10 from traversing into the optimal packing position seen in the PHAGESOL peptide against W73 and L135. In the two derivative PHAGESOL-Q8F and Q8F/L12F peptides (C and D respectively) with F located at position 8 there is minimal change in conformation observed between the two simulations and both have poor affinities for eIF4E. F8 forms favorable interactions with the surface of eIF4E and prevents any significant movement of the helix across the surface of the protein. In C) and D) both L12 (PHAGESOL-Q8F) and F12 (PHAGESOL- Q8F/L12F) pack against eIF4E with L12 making the more optimal interactions as reflected by the higher affinity of this peptide. Deviations in the planarity of the tyrosine and phenylalanine ring systems are within the tolerances of the torsional restraints of the MD simulations. [31].