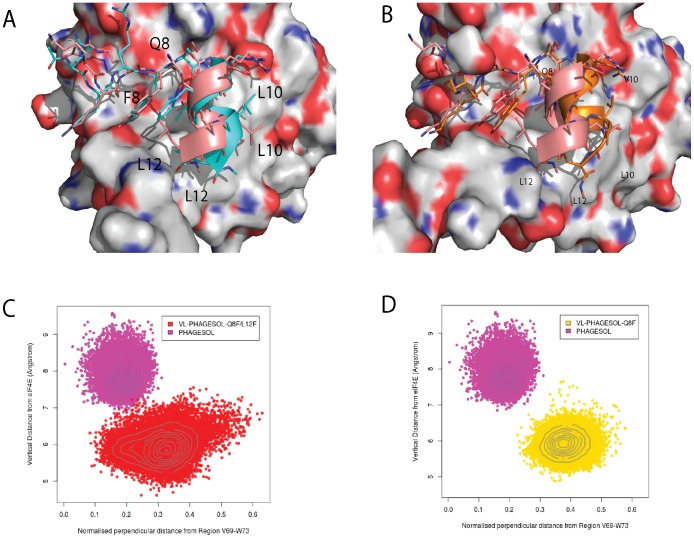
Figure 9. The precise sites of interaction for eIF4E interacting peptides are non-identical and display distinctive conformational differences.
A) Overlay of representative snapshots from the VL-PHAGESOL-Q8F (cyan) and VL-PHAGESOL (salmon) peptide simulations bound to eIF4E. The deviation in helical movement between these two peptides is dramatic with F8 in the VL-PHAGESOL-Q8F peptide forming extensive interactions with eIF4E and preventing the lateral movement across the surface of eIF4E seen for VL-PHAGESOL. The lateral movement of VL-PHAGESOL is facilitated by Q8, which forms a hydrogen bond with S5 of the bound peptide, and also forms less interactions with eIF4E. It also allows the C-terminal of the helix to unwind and allow L12 to pack optimally into the volume that F8 would occupy in the other peptide. B) Overlay of representative snapshots from the PHAGESOL and VL-PHAGESOL peptide simulations bound to eIF4E. These peptides only differ at the position 10 with PHAGESOL containing a V and VL-PHAGESOL possessing an L. The presence of Q8 allows the helix to move more independently enabling the residue at position to dictate the final packing arrangements of the peptide. V10 packs optimally in the PHAGESOL peptide against the surface of eIF4E which induces the lateral movement of the peptide. L12 can still pack optimally. L10 in the VL-PHAGESOL peptide also packs against a hydrophobic area of eIF4E, closely located to where V10 packs, but due to the greater length of the alkyl chain allows the peptide to pack in a different conformation with eIF4E. A conformation where the C-terminal end unwinds to allow L12 to pack into an alternative optimal position against eIF4E. C) and D) are plots showing the distribution of the relative positions of the helical portion of the eIF4E bound peptide with respect to the surface of the protein throughout their individual simulations. The plots show the distinct conformational differences of the peptides in their interactions with eIF4E. The PHAGESOL peptide (magenta) and VL-PHAGESOL (yellow) both have very distinct conformational populations whilst the VL-PHAGESOL-Q8F/L12F peptide (red) has a much more dispersed population that overlaps with the conformational space of the VL-PHAGESOL peptide (see figure 4S for definition of the conformational measurement made). Deviations in the planarity of the tyrosine and phenylalanine ring systems are within the tolerances of the torsional restraints of the MD simulations. [31].