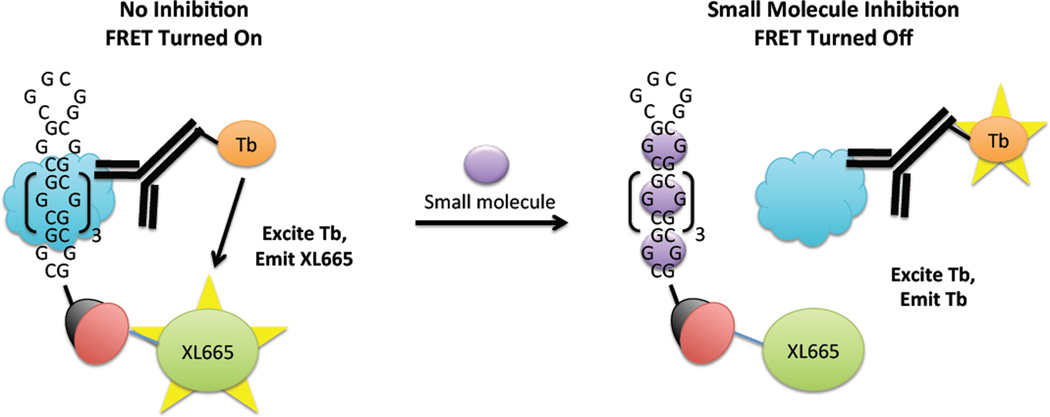
Figure 1.
Schematic of the protein displacement assay that was used to identify small molecule inhibitors of the r(CGG)12-DGCR8Δ interaction and to determine their potencies. The r(CGG)12 oligonucleotide is labeled with a 5’-biotin while DGCR8Δ (blue cloud) contains a histidine (His) tag. Left, in the absence of inhibitor, DGCR8Δ binds to r(CGG)12. Binding is quantified by using two antibodies that form a FRET pair—an anti-His antibody labeled with Tb that binds to DGCR8Δ and streptavidin labeled with XL665 that binds to r(CGG)12. The two fluorophores are within close enough proximity to form a FRET pair. Tb is excited at 345 nm; the resulting emission (~545 nm) excites XL665, which emits at 665 nm. Right, in the presence of inhibitor, the r(CGG)12-DGCR8Δ interaction is disrupted, and the two fluorophores are not within close enough proximity to form a FRET pair. Therefore, emission is only observed at 545 nm (due to Tb). XL665 emission is not observed.