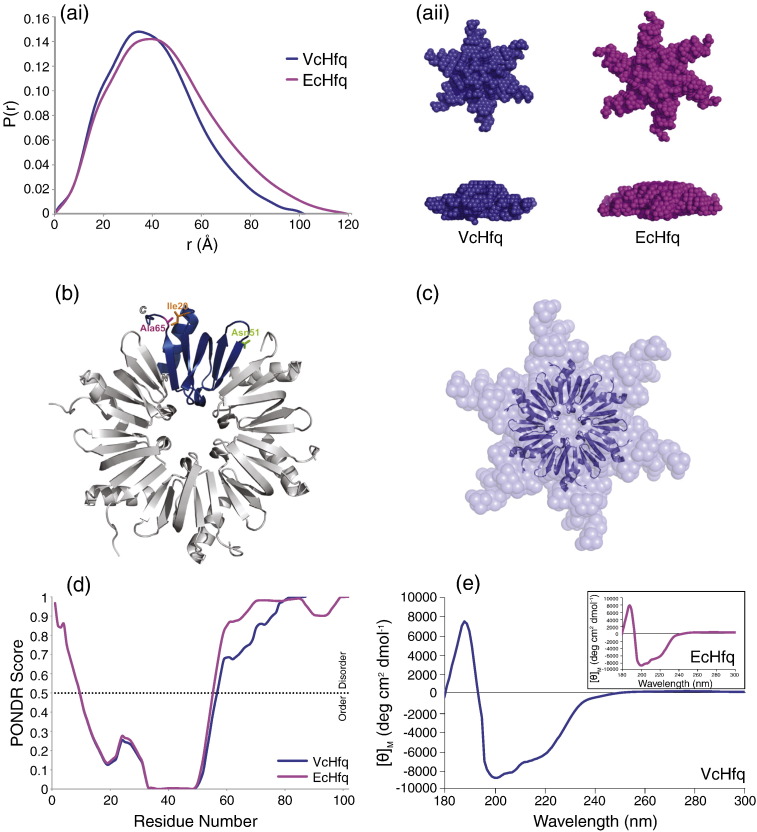
Fig. 2.
Structural studies of VcHfq and EcHfq. (a) SAXS. (i) Pair distance distribution function, P(r), plots for VcHfq (blue) and EcHfq (magenta) calculated by an indirect Fourier transformation of the SAXS data using GNOM.19 (ii) Surface representations of ab initio models for VcHfq (left; blue) and EcHfq (right; magenta) calculated using DAMMIF20 and visualized with PyMOL. Top and side views are shown for each protein. (b) Homology model of the VcHfq NTD. A ribbon representation of a homology model of the VcHfq NTD hexamer visualized with PyMOL. One monomer has been colored blue and the three positions that differ from the EcHfq NTD are highlighted on this monomer. One hundred models were calculated using MODELLER 9v821 with the structure of the EcHfq hexamer (PDB ID: 1HK9)14 as the template. The model shown is the one with the lowest discrete optimized protein energy potential. (c) Superposition of the VcHfq NTD homology model onto the ab initio model of VcHfq generated from the SAXS data. The structures were superimposed using SUPCOMB22 and visualized with PyMOL. (d) PONDR predictions for VcHfq and EcHfq. Potentially disordered regions of VcHfq (blue) and EcHfq (magenta) were identified using the PONDR VL-XT algorithm.23–25 The output from the predictor is a value between 0 and 1 with values of 0.5 and above indicating that a given amino acid is disordered. (e) CD spectra for VcHfq and EcHfq. The CD spectrum for VcHfq from 180 nm to 300 nm is shown in the main panel and the CD spectrum for EcHfq, over the same wavelength range, is shown as an inset.