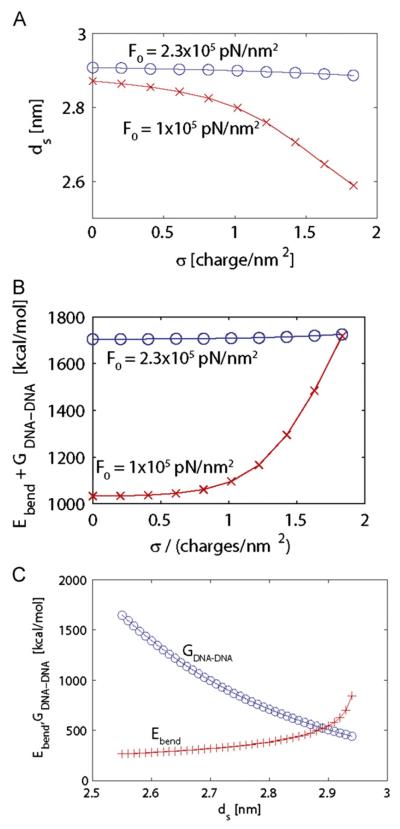
Fig. 6. Bending energy for dsDNA in an HBV core.
(A) Theoretical prediction for the dsDNA interstrand spacing ds, calculated from Eqs. (2)–(4) and Eq. (6) as a function of the C-termini charge density, σ. The last point in each curve corresponds to the charge density for Cp183. Calculated values are shown for the empirically estimated value of F0=2.3 × 105 pN/nm2 (Purohit, Kondev, and Phillips, 2003) with blue circles; for comparison, the predicted inter-strand spacing values are shown for a smaller interaction energy F0=1 × 105 pN/nm2 with red x symbols. (B) Calculated values of the destabilization energy (ΔGbend+ΔGDNA•DNA), (Eqs. (2) and (3)) for F0=2.3 × 105 pN/nm2 (blue circles) and F0=1 × 105 pN/nm2 (red × symbols). Greater values for ds are prevented due to packing constraints. (C) The bending energy ΔGbend (red + symbols) and the interaction energy ΔGDNA•DNA (blue circles) are shown as a function of ds for charge density σ 1.83 charges/nm2. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.).