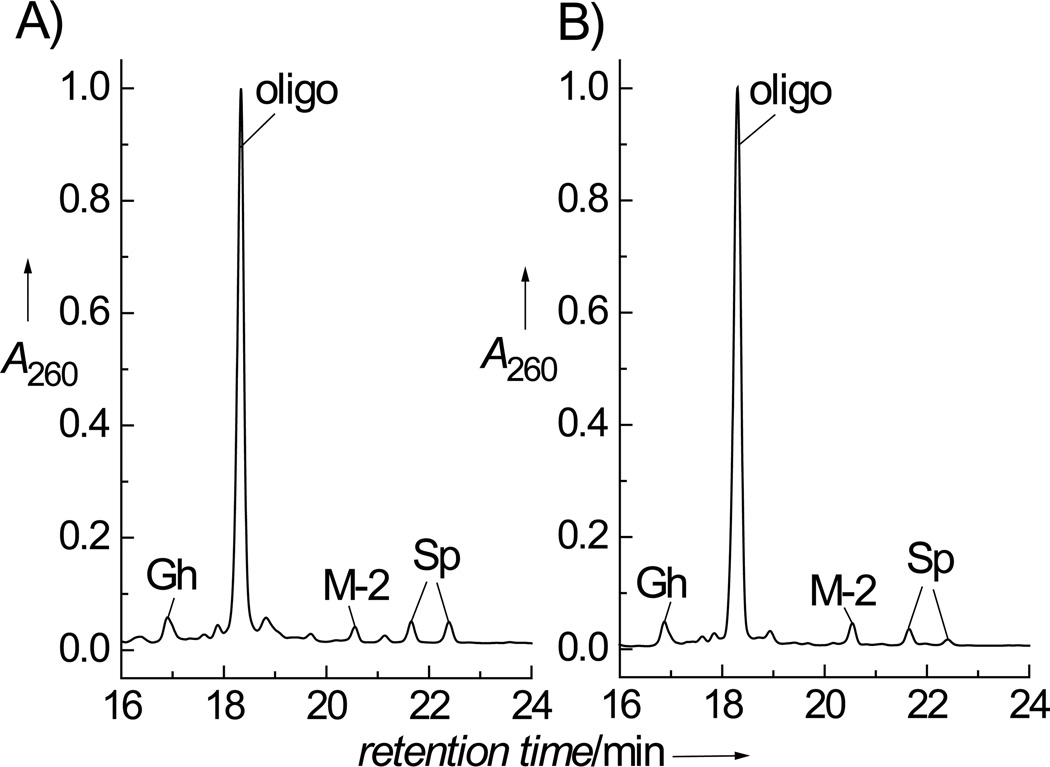
Figure 2.
Anion-exchange HPLC elution profile of the end-products derived from the oxidation of the single-stranded oligonucleotide, 5’-d(CCATCGCTACC) by CO3•− radicals. A) The 5’-d(CCATCGCTACC) sequence (0.01 mM) was irradiated for 20 s in air-equilibrated buffer solution (pH 7.5) containing 2 mM [Co(NH3)4CO3]+. B) The 5’-d(CCATCGCTACC) sequence (0.01 mM) was irradiated for 10 s in air-equilibrated buffer solution (pH 7.5) containing 300 mM NaHCO3 and 10 mM Na2S2O8. A 100 W Xe arc continuous light source was used in both cases (300 – 340 nm). HPLC elution conditions (detection at 260 nm): 10 – 90% linear gradient of solvent B (10% acetonitrile and 90% 1.5 M ammonium acetate) in solvent A (10% acetonitrile and 90% water) for 30 min at a flow rate of 1 mL/min. The fractions containing the unmodified oligonucleotide (labeled oligo), the cyclic cross-linked adduct (G*CT*), the oligonucleotides with the single G residue converted to the guanidinohydantoin lesion (Gh), and spiroiminodihydantoin lesions with (+)-R-Sp and (−)-S-Sp configurations eluting at 21.7 min, and 22.4 min, respectively, were identified as discussed in the text (the absolute configurations of the Sp lesions were determined as described by Durandin et al.;[32] note: an alternate R and S assignment was proposed by Cadet and co-workers[33]).