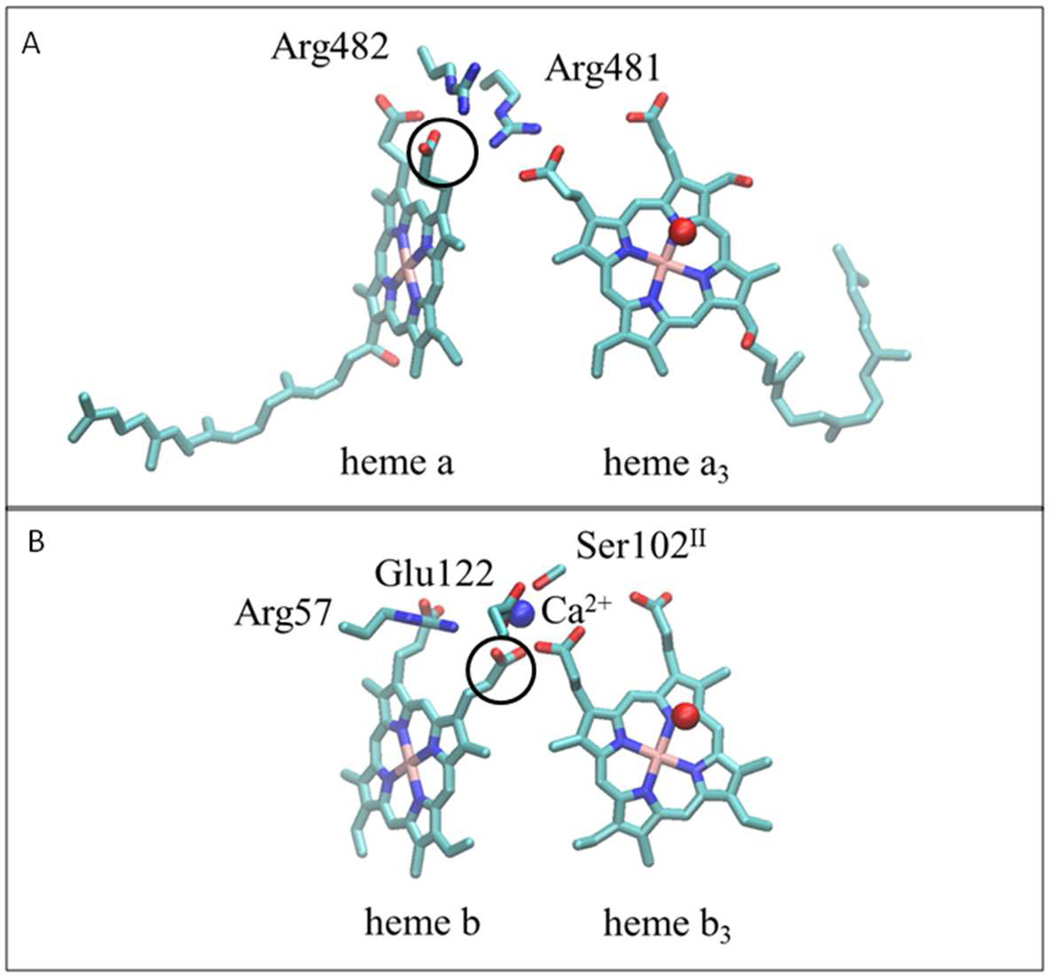
Figure 2. Comparison of heme propionate interactions in different heme-copper oxidoreductase families.
(A) In the A and B-families, represented by the structure of the A-family cytochrome c oxidase from Rhodobacter sphaeroides (PDB: 1M56), the heme propionates interact with a pair of conserved, adjacent arginine residues. The first arginine forms hydrogen bonds with propionates of the low-spin heme (heme a) and the active-site, high-spin heme (heme a3). The second arginine forms hydrogen bonds with the two propionates of the low-spin heme. (B) In the C-family oxidase from Pseudomonas stutzeri (PDB: 3MK7), the Ca+2 binding site plays an analogous role to one of the arginines (R481 in the top panel). Note the difference in the rotational configurations of the low-spin heme propionates (circled) in the two Panels. In the A and B-families the propionate is rotated out to form a hydrogen bond with the arginine, whereas in the C-family it is rotated in to ligate the Ca+2 and to allow room for the arginine to form hydrogen bonds with the low spin heme propionates. This structural feature is also found in the cNOR and qNOR families. The residue numbering in panel (B) is from the cbb3 oxygen reductase from P. stutzeri.