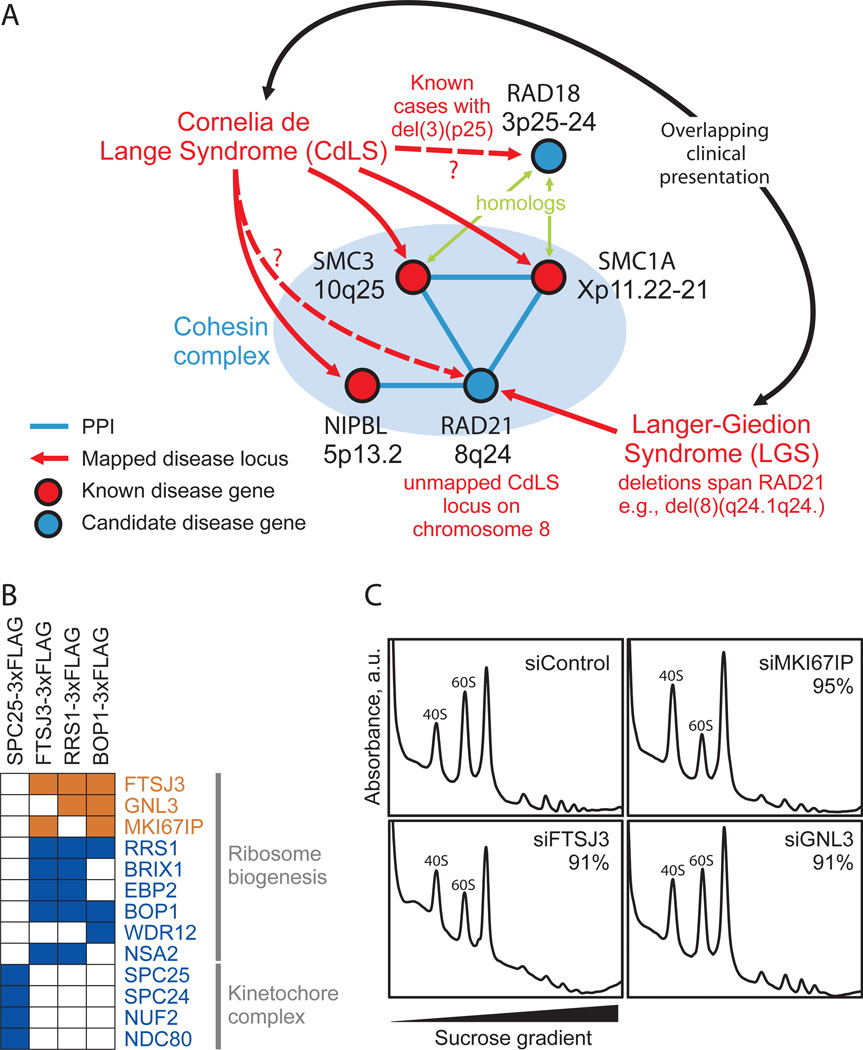
Figure 5. Membership in complexes predicts protein function and disease associations.
A- Three of four proteins mapped to the cohesin complex account for roughly half of cases of the human congenital disorder Cornelia de Lange syndrome (Pie et al., 2010), implicating the fourth component, RAD21, as a candidate disease gene. This association may explain similarities in clinical presentation between CdLS and Langer-Giedion syndrome, as the latter patients routinely harbor RAD21 deletions, e.g. (McBrien et al., 2008; Wuyts et al., 2002). B- Confirmation of ribosome biogenesis candidate (orange) associations with annotated components (blue) by AP/MS analysis of tagged proteins (top). Colored squares indicate validation (see Extended Experimental Procedures). C- Polysome profiling after siRNA targeting in tissue culture supports functional roles in ribosome biogenesis for three candidate proteins. Knockdown of MKI67IP, FTSJ3, and to a lesser extent GNL3, results in 60S ribosomal subunit biogenesis defects manifested by a reduced ratio of free 60S to 40S ribosomal subunits during gradient sedimentation as compared to control. Percentages indicate siRNA knockdown efficiency as measured by qRT-PCR.