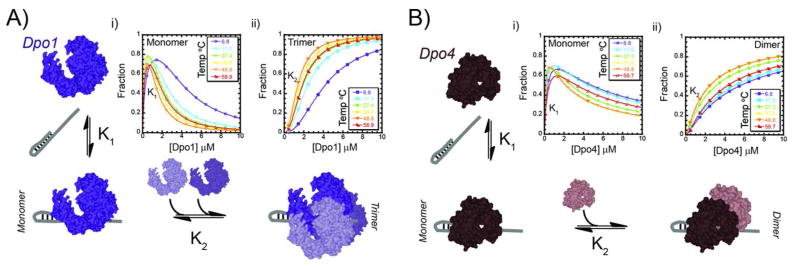
Abstract
Differentiation of binding accurate DNA replication polymerases over error prone DNA lesion bypass polymerases is essential for the proper maintenance of the genome. The hyperthermophilic archaeal organism, Sulfolobus solfataricus (Sso), contains both a B-family replication (Dpo1) and a Y-family repair (Dpo4) polymerase and serves as a model system for understanding molecular mechanisms and assemblies for DNA replication and repair protein complexes. Protein crosslinking, isothermal titration calorimetry, and analytical ultracentrifugation have confirmed a previously unrecognized dimeric Dpo4 complex bound to DNA. Binding discrimination between these polymerases on model DNA templates is complicated by the fact that multiple oligomeric species are influenced by concentration and temperature. Temperature dependent fluorescence anisotropy equilibrium binding experiments were used to separate discrete binding events for formation of trimeric Dpo1 and dimeric Dpo4 complexes on DNA. The associated equilibria are found to be temperature dependent, generally leading to improved binding at higher temperatures for both polymerases. At high temperatures, DNA binding by Dpo1 monomer is favored over Dpo4 monomer, but binding of Dpo1 trimer is even more strongly favored over Dpo4 dimer, thus providing thermodynamic selection. Greater processivities of nucleotide incorporation for trimeric Dpo1 and dimeric Dpo4 are also observed at higher temperatures, providing biochemical validation for the influence of tightly bound oligomeric polymerases. These results separate, quantify, and confirm individual and sequential processes leading to formation of oligomeric Dpo1 and Dpo4 assemblies on DNA and provide for a concentration and temperature dependent discrimination of binding undamaged DNA templates at physiological temperatures.
Keywords: DNA replication, polymerase, thermodynamics, Dpo1, Dpo4, archaea, DNA binding
INTRODUCTION
Over the past two decades, a multitude of DNA polymerases have been discovered and classified into at least six separate families (1). Many organisms have multiple DNA polymerases with humans having fifteen (2). Most traditional B-family DNA polymerases are involved in faithful copying of our genome, while Y-family DNA polymerases have more flexible active sites allowing for synthesis across locations of DNA damage in an effort to maintain uninterrupted DNA synthesis during replication. Binding and recognition of normal or damaged DNA bases require that each DNA polymerase has a precise specificity with the appropriate DNA template to maintain fidelity of replication or repair directed by interacting proteins at the replication fork. Specificity is increased through interactions with shared accessory proteins for DNA replication and repair polymerases at the site of catalysis. Alternatively, the repeated shuttling between polymerization and exonuclease states of B-family DNA polymerases at sites of damage (3) may locally destabilize binding, allowing a Y-family polymerase to bind more specifically to bypass the lesion. Therefore, a question arises as to how each polymerase is regulated with regards to binding the correct DNA substrate.
DNA polymerases Klenow (4), T4 gp43 (5), human pol β (6, 7), and African swine fever virus polymerase X (8) have all been found to form 2:1 complexes with DNA (9). Other DNA polymerases appear at the replication fork as multimers during DNA replication through interactions with their accessory proteins (10). Interestingly, DNA replication polymerases have also been found to exchange freely from solution during active replication, suggesting that either direct interactions between polymerases or indirect interactions through accessory proteins provide mechanisms for switching enzymes at the site of catalysis (11, 12). Therefore, it is likely that the high concentration of DNA polymerases within or around replisome complexes is a common mechanism for coordinated DNA synthesis, increased kinetics, and coupled replication and repair to maintain the genomic integrity of the cell.
DNA replication in archaea serves as an important and relevant model system for detailing the molecular mechanisms of DNA polymerases homologous to their eukaryotic counterparts (13–15). Contained within the archaeal Sulfolobus solfataricus genome are DNA polymerases from both the B-family replication (Dpo1) and Y-family lesion bypass (Dpo4) families. Both individual DNA polymerases have been structurally characterized (16–18), have similar specificities for DNA (19, 20), and robust kinetics (21–23), but differ in their fidelities for nucleotide incorporations (21, 24–26). Dpo4’s lower fidelity, as well as the ability for Sulfolobus to survive in the absence of this protein (27), highlights a nonessential role in DNA replication. Direct interaction between Dpo1 and Dpo4 has also been noted and is thought to be important for uninterrupted lesion bypass during DNA replication (28). We have also shown that Dpo1 can form a trimeric complex in the presence of DNA (20), suggesting that homo and heteroligomeric DNA polymerase complexes exist.
Quantification of binding various DNA polymerases to DNA has been examined using a number of techniques to characterize this single binding event. The resulting temperature dependent thermodynamic binding parameters can provide insight into the specificity of the binding process through determination of the heat capacity change (ΔCop) (29, 30). Although a strongly negative ΔCop has been shown to be the thermodynamic signature of sequence-specific binding (30), the non-sequence specific binding to primer template DNA by the A-family DNA polymerases from Thermus aquaticus (Taq) (31) and Escherichia coli (Klenow) (32) is also associated with large and negative ΔCop values (29, 33–35). Even though there is no sequence specificity, the negative ΔCop is consistent with the high structural complementarity (29) of the DNA polymerase binding to the primer template junction, visualized in a variety of crystal structures (18, 36, 37). The inherent thermostability of proteins from Sso (where the growth temperature is ~75 °C) allows us to fully investigate the energetic constraints of DNA polymerase binding to DNA. Access to this broad temperature range results in a more complete thermodynamic characterization of the differences in binding B and Y-family polymerases to an undamaged DNA primer-template. These thermodynamic differences can be evaluated directly by determining the affinities (Kd), free energies (ΔGo), heat capacity changes (ΔCop), and stoichiometries (n) for binding each polymerase.
Here, we use chemical crosslinking, isothermal calorimetry (ITC), and analytical ultracentrifugation (AUC) to show that Dpo4, like Dpo1, can also form an oligomeric complex on DNA. Using AUC, we have found both a strong concentration dependent and modest temperature dependent shift in the reaction boundaries, highlighting changes in Dpo4 monomer-dimer and Dpo1 monomer-trimer equilibria. Temperature dependent equilibrium fluorescent anisotropy binding experiments were used to separate the free energy (ΔGo) of binding either monomer or higher order oligomeric DNA polymerases (Dpo1 or Dpo4) states. For both polymerases, we have detected an initial higher affinity monomeric binding site followed by sequential binding of additional polymerase molecules to form either trimeric Dpo1 or dimeric Dpo4 complexes on DNA.
Separation and quantification of these individual binding events reveal that a Dpo1 monomer binds to DNA with only slightly greater affinity than Dpo4 up to 50 °C. This binding affinity difference is exaggerated at the highest temperatures, suggesting that binding of Dpo1 to undamaged DNA templates is favored at physiological growth conditions for Sso. The free energy associated with trimer Dpo1 binding to DNA is significantly more favorable than that associated with dimer Dpo4 DNA binding, and this difference increases strongly with increasing temperature. Enzymatic evidence showing greater processivities for Dpo1 and Dpo4 at higher temperatures and protein concentrations is used to explain the role of temperature and oligomeric state in promoting DNA polymerase assembly, stability, and kinetics at the replication fork. Collectively our results indicate that the binding specificities of multiple oligomeric archaeal DNA polymerases are regulated by changes in cellular concentrations and temperature for efficient DNA binding recognition and synthesis.
EXPERIMENTAL PROCEEDURES
Materials
Oligonucleotide substrates including the 37 nucleotide (nt) DNA hairpin, 5’-fluorescein or 5’-Cy3 labeled DNA were purchased from Integrated DNA technologies (IDT, Coralville, IA). The sequence of the 37 nucleotide DNA hairpin was 5’-TTTTTTTTTTCCCGGGCCGGCGTTTCGCCGGCCCGGG, which included a 12 base-pair duplex region, a three residue loop, and a ten residue single strand template. DNA was dissolved in annealing buffer [20 mM HEPES (pH 7) and 200 mM NaCl], heated to 95 °C for 15 minutes, and then cooled to room temperature by turning off the hot plate overnight. M13mp18 was purchased from USB Corporation (Cleveland, OH). All radiochemicals were purchased from MP Biochemicals (Santa Ana, CA) or Perkin Elmer (Waltham, MA). Commercial enzymes were from NEB (Ipswich, MA). All other chemicals were analytical grade or better.
Dpo1 and Dpo4 were purified as described (17, 20) except for a few modifications. The exonuclease deficient version of Dpo1 (D231A/D318A) was recloned into pET30a (NdeI/XhoI) to introduce a stop codon and remove the C-terminal His tag. Both polymerases were expressed using an autoinduction protocol (38) using Rosetta 2 cells (Stratagene). Cells were lysed by sonication and heat treated at 70 °C for 30 minutes followed by centrifugation. The Dpo1 lysate was purified using a HiTrap MonoQ, heparin, and Superdex-200 gel filtration columns. The wild-type untagged Dpo4 lysate was purified using a HiTrap MonoQ, heparin, and hydroxylapatite (Sigma-Aldrich) columns (17).
Crosslinking Studies
Dpo4 was dialyzed in crosslinking buffer [50 mM HEPES (pH 7.0), 150 mM NaCl, 10 mM EDTA] and reduced using 10 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP-HCl) (Thermo Scientific, Rockford, IL). Dpo4 (8 LM) was then either incubated alone or with 10 μM DNA 37 nucleotide hairpin for 5 minutes at room temperature. Chemical crosslinking experiments were initiated with 0.33 mM 1,11-bis-maleimidotriethyleneglycol [BM(PEG)3] or ethylene glycol bis[succinimidylsuccinate] [EGS] (Pierce, Rockford, IL) targeting free cysteines or free amines, respectively, in close proximity. The reaction was then incubated for 15 minutes at 22 °C. Products were separated on an 8% SDS-PAGE gel and stained with coomassie dye.
Isothermal Titration Calorimetry (ITC)
Prior to analysis, titrants and analytes were dialyzed against Buffer A [20 mM HEPES-NaOH (pH 7), 150 mM NaCl, 5% Glycerol, 10 mM Mg(OAc)2, 0.2 mM DTT], filtered by centrifuge tube filters (0.22 μM, SPIN-X, Corning Inc., NY), and degassed. Isothermal titration calorimetry was performed using a VP-ITC (MicroCal Inc., Northampton, MA) as described previously (20). Titrations were performed by titrating 400 μM 37-nt hairpin (primer template) (5–8 μL aliquots) into 25 μM Dpo4 at 15 oC or 60 °C. The heats of the reaction were corrected for the heat of dilution by subtracting the signal after reaching saturation. All data were fit using Origin 7.0 (MicroCal) according to the following equation:
| (1) |
where V0 is the volume of the cell, ΔHo is the enthalpy of binding per mole of ligand, [M]t is the concentration of DNA including both bound and free fractions, Ka is the association constant, [L]t is the total ligand (Dpo4) concentration, and n is the stoichiometry of the reaction (20, 39).
Analytical ultracentrifugation (AUC)-Sedimentation velocity experiments
Sedimentation velocity experiments were performed using an Optima XLI analytical ultracentrifuge (Beckman Coulter, Fullerton, CA) equipped with a prototype fluorescence optical system (Aviv Biomedical). Samples of protein alone or with (50 nM) fluorescein labeled 37-nt hairpin (primer-template) DNA in Buffer A were loaded into ultracentrifuge cells at various concentrations (0, 10, 50, 100, 200, 500, 1000, 2000, 5000, and 10000 nM) into a double-sector charcoal-epon centerpiece in either a 4- or 8-hole titanium rotor and subjected to an angular velocity of 45,000 rpm with the temperature at 10, 20, or 30 °C. Absorbance or fluorescence scans as a function of radial position were collected by scanning at 280 nm (protein alone) or at 488 nm (protein with fluorescein DNA) at 20-μm radial increments, averaging 3 revolutions/scan. Sedimentation velocity boundaries were analyzed in the least squares sedimentation coefficient distribution (ls-g*(s)) model using program SEDFIT (version 12.1) (40). The sedimentation coefficient, s, is given by Svedberg’s equation:
| (2) |
where MW is the molecular weight, Dt is the diffusion coefficient, ῡ is the partial specific volume, ρ is the solvent density, R is the universal gas constant, and T is temperature. Observed weight average sedimentation coefficients were converted to s20,w (standard conditions of 20 °C in water) accounting for partial specific volumes, buffer densities, and viscosities, calculated using SEDNTERP (41, 42).
Fluorescence anisotropy, equilibrium binding, and thermodynamic parameters
A 5’ Cy3-labeled 37-nt hairpin primer template DNA construct, described previously (20), was used in the fluorescence anisotropy experiments. Titrations were performed in Buffer A using a fixed concentration of DNA (4 nM) and varying concentrations of protein (0–20 LM). Anisotropy measurements were performed using a FluoroMax-3 spectrofluorimeter (HORIBA Jobin Yvon) equipped with automated polarizers and regulated with a thermostated cuvette holder. The DNA and protein were allowed to equilibrate at each temperature for at least 30 minutes prior to measuring the anisotropy. Titrations at higher temperatures (>45 °C) were performed in a capped cuvette to limit concentration changes due to evaporation. Fluorescence was excited at 550 nm, and the emission with various combinations of polarizers was monitored at 564 nm during a 5 sec integration time. The fluorescence anisotropy, r, was calculated automatically by the instrumental software using the equation:
| (3) |
where I is the polarized fluorescence intensity with subscripts V and H identifying either vertical or horizontal polarized light, respectively. The G-factor is a correction for the difference in sensitivities of detection for horizontal and vertically polarized light. In all titrations, protein was titrated into DNA. After each addition, the protein was equilibrated until no further change in anisotropy was detected, generally 1 minute. The fluorescence intensity of Cy3 is known to change with temperature (43). Therefore, the slits were adjusted accordingly at each temperature to give a total fluorescence signal of approximately 106 counts per second (CPS) for each titration. As a control for specific binding, the absolute fluorescence intensity at 564 nm did not change significantly with addition of a high concentration of either Dpo1 or Dpo4 measured at the beginning and end of the experiment.
Anisotropy data were fit to either a single
| (4) |
or sequential binding sites equation
| (5) |
where A is the change in anisotropy, P is either Dpo1 or Dpo4 concentration, Kd is the dissociation constant for each binding event (subscript 1 or 2), and n is the stoichiometry (2 for Dpo1 and 1 for Dpo4). At least three independent titrations were performed at each temperature to obtain average Kd1 and Kd2 parameters. Kd1 and Kd2 were used to directly calculate free energy change (ΔGo) for monomer:
| (6) |
and oilgomer (Dpo1 trimer or Dpo4 dimer):
| (7) |
where R is the universal gas constant and T is temperature.
Thermodynamic parameters were extracted from the temperature dependence of the Gibbs-Helmholtz plot from a multiparametric fit according to the following equations
| (8) |
| (9) |
| (10) |
where ΔGo is the standard free energy change, ΔHo is the change in enthalpy, and ΔSo is the change in entropy, using a constant heat capacity ( ) at each temperature, T. TH is the temperature in which ΔHo = 0, TS is the temperature where TΔSo = 0.
The binding data were also fit to a van’t Hoff plot of lnKapp versus 1000/T according to the following equation
| (11) |
where Kapp is the apparent equilibrium constant and R is the universal gas constant.
Buried Surface Area Calculations
Solvent accessible surface areas for Dpo4 bound to DNA (PDB ID: 2RDJ) (44) were calculated for buried nonpolar (ΔAnp) and polar (ΔAp) surface areas using a 1.4 Å probe radius as described (45). The heat capacity change associated with binding was calculated from a surface area-based model according to Spolar et al (46).
| (12) |
Polymerase/DNA binding simulations
The cumulative binding data for Dpo1 and Dpo4 were fit and modeled according to the minimal kinetic scheme outlined in Figure 6A & B using a simulation with Berkeley Madonna (University of California, Berkeley) (see Supporting Information for a full description of the parameters and equations used).
Figure 6.
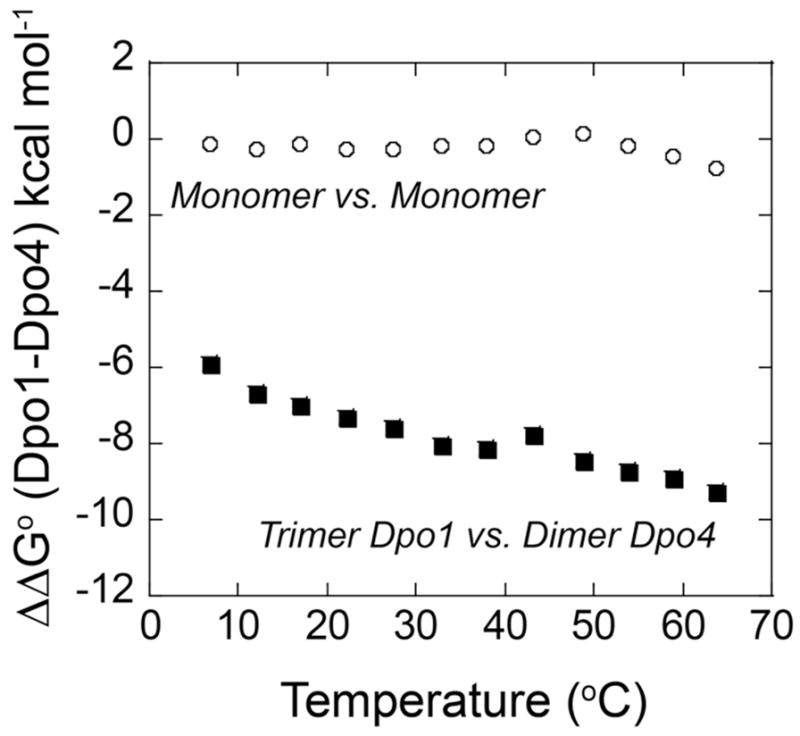
Gibbs free energy differences (ΔΔGo) between Dpo1 and Dpo4 monomers (○) or between trimeric Dpo1 and dimeric Dpo4 (■) plotted as a function of temperature.
DNA polymerase processivity
Processivity experiments for Dpo1 and Dpo4 were performed and analyzed as previously described (20) but at additional temperatures. 5’-32P end-labeling of a DNA primer was performed using Optikinase (USB) and 32P γ-ATP according to manufacturer’s directions. Primed M13mp18 DNA template (40 nM) was preincubated with various concentrations of Dpo1 or Dpo4 (as indicated in the Figure legends) at the experimental temperatures in Buffer A, and the reaction was initiated with the addition of dNTPs (0.1 mM each) and/or 30 μg single strand salmon sperm DNA (Invitrogen, Carlsbad, CA). One volume of stop solution [50 mM NaOH, 1 mM EDTA, 3% (w/v) Ficoll (Type 400, Pharmacia), 0.05 % (w/v) bromocresol green, 0.04 % (w/v) xylene cyanol] was added to terminate the processivity reactions after 60 minutes for 40 °C, 30 minutes for 50 °C, and 10 minutes for 60 or 70 °C reactions. Aliquots were run on an alkaline agarose gel (0.8% agarose, 50 mM NaOH, 1 mM EDTA) or denaturing 20% PAGE gel, dried and imaged using a Storm Phosphorimager (GE Healthsciences). Quantification of the mean band lengths was performed using ImageQuant software (v5.0) compared with a 32P end labeled 1kb ladder (Invitrogen).
RESULTS
Detection of dimeric Dpo4
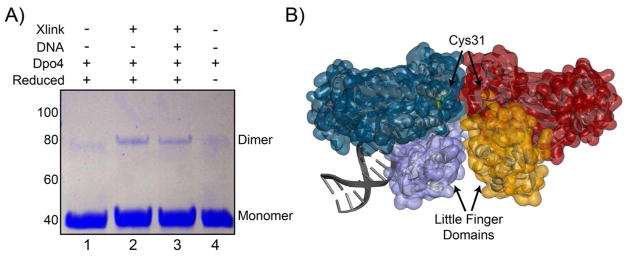
After purifying Dpo4, we noticed a small amount of protein consistent with a covalent dimer on SDS-PAGE gels, especially under non-reducing conditions. We investigated the validity of a possible Dpo4 dimer using protein cross-linking. Chemical crosslinkers, BM(PEG)3, which targets native reduced cysteines, or EGS, which targets free amines in close proximity (<18 Å), were both used to capture a dimer in solution. Dpo4 contains a single native cysteine residue (C31) which allows for the possibility of inferring information about the structure of a crosslinked species. Using the thiol-thiol crosslinker [BM(PEG3)], we were able to crosslink a dimeric Dpo4 both in the absence and presence of DNA (Figure 1A, lanes 2 and 3). There is no significant difference in the amount of crosslinked Dpo4 complex in the presence of DNA. Therefore, a monomer-dimer equilibrium exists both on and off DNA. Unreduced Dpo4 loaded onto the SDS-PAGE gel also shows a small quantity of dimeric product suggesting that a disulfide bond can form between subunits without added crosslinker (Figure 1, lane 4). Reduction of this disulfide bond with TCEP reduces the fraction of dimer in favor of monomer (Figure 1, lane 1). We were also able to detect an equivalent reduced dimeric Dpo4 species using an amino-amino crosslinker (EGS) which crosslinks lysine residues in an interface (data not shown). These crosslinking results suggest that dimeric Dpo4 complexes can exist in solution and that at least one conformation positions the single cysteine residue at the protein-protein interface, similar to that observed in an x-ray structure of Dpo4 bound to DNA (Figure 1B) (47).
Figure 1.
Dimeric Dpo4 complex formation. A) Covalent protein crosslinking of Dpo4 in the absence and presence of DNA hairpin or thiol-thiol crosslinker [BM(PEG)3]. Lane 1: reduced Dpo4, Lane 2: reduced Dpo4 with crosslinker, Lane 3: reduced Dpo4-DNA complex (37-nt hairpin) with crosslinker, and Lane 4: unreduced Dpo4. The positions corresponding to monomer (40 kDa) and dimer (80 kDa) forms of Dpo4 are shown in the right margin. B) X-ray structure of one possible dimeric Dpo4 conformation found in the crystal unit (PDB ID: 2W9B) consistent with crosslinking at the C31 interface between molecules. Highlighted in purple and orange surfaces are the little finger domains from each Dpo4 molecule.
Stoichiometry of Dpo4 binding to DNA by isothermal titration calorimetry
In order to verify that a dimeric Dpo4 complex can exist on DNA over a broad temperature range, we used ITC to quantify the thermodynamics and stoichiometry of binding at 15 °C and 60 °C (Figure 2). At 15 °C, binding is primarily entropically driven, and a fit of the binding isotherm to Equation 1 gave the following values: Kapp = 6.5 ± 0.6 x 105 M−1, ΔHoITC = 8.1 ± 0.2 kcal mol−1, and n = 0.64 ± 0.01. The resulting ΔGoITC is −7.7 kcal mol−1 and the calculated entropic contribution (TΔSITC) is 15.7 kcal mol−1. At 60 °C the binding is primarily enthalpically driven, and a fit of the binding isotherm to Equation 1 gave Kapp = 2.2 ± 0.5 x 106 M−1, ΔHoITC = −8.0 ± 0.5 kcal mol−1, and n = 0.66 ± 0.01. The resulting ΔGoITC is −9.7 kcal mol−1 and the calculated TΔSITC is 1.7 kcal mol−1. Importantly, the stoichiometries at 15 °C and 60 °C are similar and are consistent with more than one molecule of Dpo4 bound to DNA. A dimer was also seen using chemical crosslinking. Although the titrations appear to go to completion, the stoichiometries do not saturate to n = 0.5 (two Dpo4 molecules per DNA) indicating that the dimeric Dpo4 complex is in equilibrium with monomer-DNA complex under these conditions.
Figure 2.
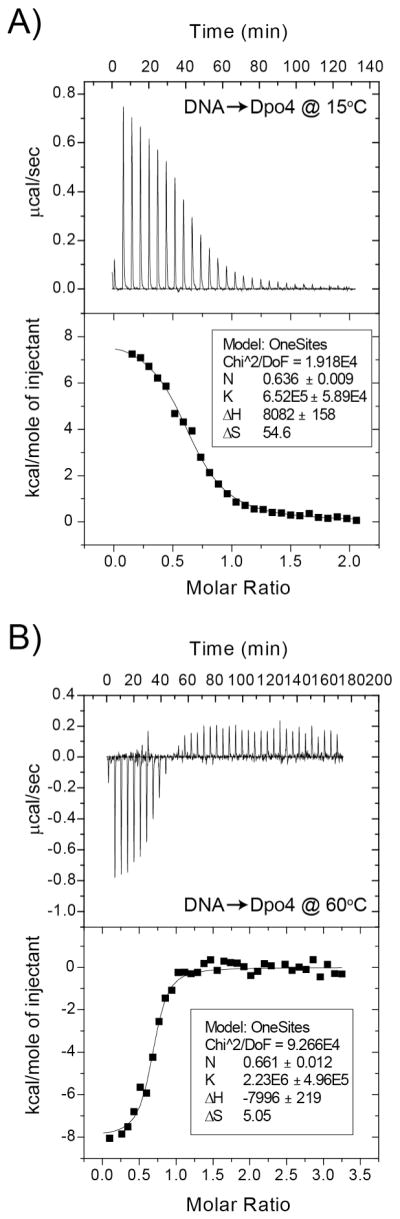
Stoichiometry and thermodynamics of Dpo4 binding to DNA. ITC titration of 400 μM DNA hairpin into 25 μM SsoDpo4 at A) 15 °C and B) 60 °C as described in Materials and Methods. Data were fit using Equation 1 to yield stoichiometries (n) 0.64 ± 0.01 or 0.66 ± 0.01 (DNA:Dpo4), apparent equilibrium association constants (Kapp) 6.52 ± 0.59 x 105 or 2.23 ± 0.50 x 106 M, enthalpy changes (ΔH°ITC) 8.08 ± 0.16 or 8.00 ± 0.22 kcal mol−1, and entropy changes (ΔSoITC) 54.6 or 5.1 cal mol−1 K−1 at 15 °C and 60 °C, respectively.
Analytical velocity sedimentation detects the formation of oligomeric Dpo1 and Dpo4 complexes with DNA
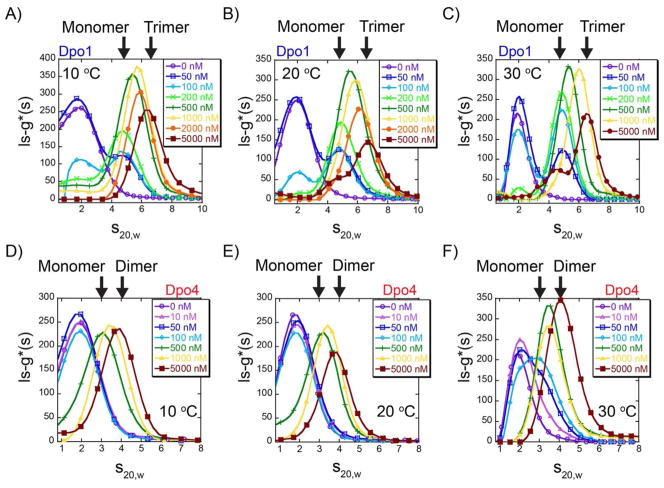
Analytical ultracentrifugation (AUC) sedimentation velocity experiments were performed with either protein alone (Dpo1 or Dpo4) at 10, 20 or 30 °C to monitor the concentration and any temperature dependent equilibria. For either Dpo1 or Dpo4 alone, there was an increase in the peak position of the sedimentation reaction boundary, ls-g*(s), with concentration (1 or 10 μM), as expected at each temperature based on 280 nm detection. The sedimentation reaction boundaries were corrected for changes in the diffusion coefficient with temperature to give the sedimentation coefficient, s20,w, and represent solution equilibrium distribution values for each experimental condition. Increasing the concentration of Dpo4 from 1 to 10 μM did not change the s20,w values significantly. At constant concentrations of Dpo1 or Dpo4, the weight average s20,w value shifts slightly larger with increasing temperature (Figure S1 of Supporting Information). Analysis of the reaction boundaries for 10 μM Dpo1 resulted in weight average s20,w distribution values of 4.19 ± 0.01, 4.29 ± 0.01, and 4.32 ± 0.01 for 10, 20, and 30 °C, respectively. Similarly, analysis of the reaction boundaries for 10 μM Dpo4 resulted in weight average s20,w distribution values of 2.52 ± 0.01, 2.55 ± 0.01, and 2.59 ± 0.01 for 10, 20, and 30 °C, respectively. Increasing s20,w values are consistent with a shift in the equilibrium towards formation of larger species. For protein alone (Dpo1 or Dpo4), these changes in s20,w are only slightly significant over this limited temperature range.
More specific information on complex assembly can be obtained by examining the reaction boundaries for titration of each polymerase with a constant concentration (50 nM) of fluorescent hairpin primer-template DNA using analytical ultracentrifugation fluorescence-detected sedimentation (AU-FDS) (48). By monitoring the reaction boundaries of fluorescent DNA in the AUC velocity experiments, we are able to examine a much greater dynamic range of protein concentrations (50 – 5000 nM) than with absorbance alone. Titration of Dpo1 at 10, 20, and 30 °C shows a clear increase in the s20,w distributions and boundaries with concentration, consistent with the detection of multiple protein bound DNA complexes (Figure 3 A–C). Here, discrete s20,w populations for monomeric and trimeric Dpo1 can be identified. Interestingly, the s20,w reaction boundaries at identical concentrations of Dpo1 bound to DNA shift towards larger species with increasing temperature more significantly than for the free protein alone. For example, specifically examining 100 nM Dpo1 across the three temperatures, the weight average s20,w values increase with increasing temperature: 4.66 ± 0.03, 4.77 ± 0.01, to 4.86 ± 0.01 at 10, 20, and 30 °C, respectively (Figure S2A of Supporting Information). At 1 μM Dpo1, the weight average s20,w values increase from 5.71 ± 0.01, 5.89 ± 0.01, to 6.04 ± 0.01 at the same temperatures, respectively (Figure 2B of Supporting Information).
Figure 3.
Solution assembly of oligomeric polymerases on DNA. Analytical ultracentrifugation velocity fluorescence detected sedimentation (AU-FDS) experiments showing the ls-g*(s) distribution profiles as a function of Dpo1 or Dpo4 concentrations: 0 (-○-, purple) , 10 (-△-, pink), 50 (-□-, blue), 100 (-◇-, cyan), 200 (-x-, light green), 500 (-+-, dark green), 1000 (-▲-, yellow), 2000 (-●-, orange) and 5000 nM (-■-, brown) at A) and D) 10 °C , B) and E) 20 °C , or C) and F) 30 °C, respectively. Every fifth data point is indicated for simplicity and all data were fit as described in Materials and Methods. s20,w positions representing monomer or trimer Dpo1 and monomer or dimer Dpo4 are indicated by arrows.
Titration of Dpo4 on fluorescent hairpin primer-template DNA using AU-FDS also shows an increase in the s20,w boundaries at each temperature (Figure 3 D–F) consistent with populations consisting of both monomeric and dimeric Dpo4-DNA complexes. Binding of Dpo4 to DNA does not appear to occur until the Dpo4 concentration exceeds 100 nM at 10, 20, or 30 °C. Moreover, the reaction boundary continually shifts to larger species between 500 – 5000 nM. Similar to the behavior of Dpo1, the reaction boundaries at constant concentrations of Dpo4 measured at 10, 20, and 30 °C also increase or shift to slightly larger weight-average complexes more significantly than protein alone. Examination of 5 μM Dpo4 across the three temperature ranges (10, 20, and 30 °C) shows that the weight average s20,w values increase from 3.79 ± 0.02, 3.84 ± 0.01, to 4.12 ± 0.01, respectively (Figure S2D of Supporting Information). The equilibrium shift in the sedimentation coefficients with temperature can be most clearly seen at 100 nM Dpo4 (Figure S2C of Supporting Information) where initial binding is only observed at 30 °C. This observation agrees well with the fluorescence anisotropy data below which measures a Kd1 value of 0.130 ± 0.004 LM at 33 °C but has larger Kd1 values at lower temperatures (Table 1).
Table 1.
Equilibrium fluorescence anisotropy binding parameters for polymerase binding to DNA
| Dpo1 | Dpo4 | ||||
|---|---|---|---|---|---|
| Temp (oC) | Kd1 (μM)a | Kd2 (μM)a | Temp (oC) | Kd1 (μM)a | Kd2 (μM)a |
| 6.8 | 0.322 ± 0.023 | 17.2 ± 0.3 | 6.8 | 0.435 ± 0.116 | 9.32 ± 0.88 |
| 12.0 | 0.208 ± 0.057 | 8.01 ± 1.21 | 12.0 | 0.329 ± 0.107 | 5.16 ± 0.05 |
| 17.0 | 0.170 ± 0.023 | 5.78 ± 0.13 | 17.0 | 0.214 ± 0.003 | 4.70 ± 1.01 |
| 22.2 | 0.109 ± 0.016 | 4.70 ± 0.78 | 22.1 | 0.176 ± 0.010 | 3.22 ± 0.72 |
| 27.4 | 0.105 ± 0.003 | 3.70 ± 0.32 | 27.3 | 0.164 ± 0.009 | 2.82 ± 0.35 |
| 32.8 | 0.094 ± 0.001 | 2.68 ± 0.11 | 32.9 | 0.130 ± 0.004 | 3.01 ± 0.92 |
| 38.0 | 0.097 ± 0.003 | 2.48 ± 0.06 | 37.8 | 0.140 ± 0.066 | 2.33 ± 0.40 |
| 43.3 | 0.144 ± 0.003 | 2.60 ± 0.22 | 43.2 | 0.133 ± 0.026 | 1.72 ± 0.46 |
| 48.8 | 0.144 ± 0.015 | 1.82 ± 0.36 | 48.8 | 0.125 ± 0.051 | 2.32 ± 0.95 |
| 53.9 | 0.113 ± 0.032 | 2.41 ± 0.79 | 53.8 | 0.155 ± 0.065 | 2.71 ± 0.99 |
| 58.9 | 0.208 ± 0.022 | 3.21 ± 0.21 | 58.9 | 0.444 ± 0.115 | 3.81 ± 0.28 |
| 63.7 | 0.198 ± 0.026 | 4.52 ± 0.40 | 65.2 | 0.674 ± 0.080 | 7.18 ± 0.93 |
Kd1 and Kd2 are the equilibrium dissociation constants for the first and second binding events, respectively. Values are means and standard errors from parameters fit to Equation 5 from at least three independent titration experiments at each temperature.
Temperature dependent separation of polymerase binding events using fluorescence anisotropy
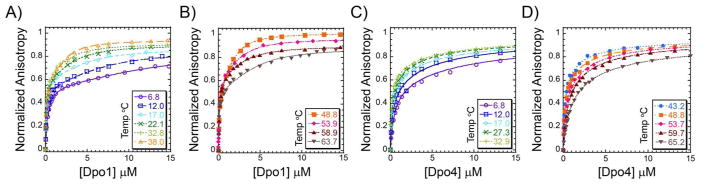
In order to investigate further the individual binding events of Dpo1 or Dpo4 on DNA, we used fluorescence anisotropy to evaluate the constants for the monomeric (Kd1) and subsequent oligomeric (Kd2) binding steps over a range of temperatures. The melting temperature (TM) of the DNA primer template hairpin was measured from a shift in the UV absorbance and found to be 88 °C (data not shown), which is well above our experimental temperature range. Protein was titrated into low concentrations of a Cy3 labeled DNA primer template hairpin and the increase in fluorescence anisotropy due to binding was monitored (Figure 4). The increase in anisotropy as a function of Dpo1 concentration was fit to either a single (Equation 4) or a sequential binding (Equation 5) model (Figure 4 A&B and Figure S3A of Supporting Information). The second and third individual binding events for Dpo1 cannot be separated from our EMSA or ITC data (20) (and manuscript in preparation); therefore, simultaneous or cooperative binding is hypothesized. The increase in anisotropy as a function of Dpo4 concentration was also fit to either Equation 4 or 5. Equation 5 provided the best fit of the data for Dpo4 (Figure 4 C&D and Figure S3B of Supporting Information). The fits to these individual equations to the data are consistent with stoichiometric values and processes measured by ITC, chemical crosslinking, AUC, EMSA, and gel filtration (20). The differences in the individual Kd values (1 & 2) for each DNA polymerase are greater than 10-fold (Table 1) and generally decrease concurrently with temperature up to 50 °C. Kd2, fit from the anisotropy experiments, is the intrinsic constant for binding of one of the two monomers in the second step, while Kd22 represents the constant for simultaneous binding of both Dpo1 monomers in the second “step” to form the trimeric Dpo1-DNA complex.
Figure 4.
Quantification of individual binding steps leading to oligomeric polymerase-DNA complexes. Representative normalized individual equilibrium fluorescence anisotropy titrations for A) - B) Dpo1 and C) - D) Dpo4 binding to DNA at low or high temperatures, respectively. Data are included for both lower 6.8 (-○-, purple), 12.0 (-□-, blue), 17.0 (-◇-, cyan), 22.1 or 27.3 (-x-, dark green), 32.8 or 32.9 (-+-, light green), 38.0 (-△-, light orange) and upper temperatures 43.3 (-●-, blue), 48.8 (-■-, dark orange), 53.7 or 53.9 (-◆-, pink), 58.9 or 59.7 (-▲-, brown), and 63.7 or 65.7 °C (-▼-, grey). The individual data points were fit to Equation 5 to determine Kd1 and Kd2 values for Dpo4 or Dpo1, respectively. At least three independent titrations were performed and fit at each temperature and the resulting values are reported in Table 1.
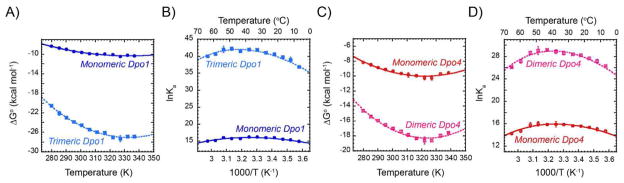
The temperature dependences of the monomeric and trimeric binding equilibria for Dpo1 to DNA are plotted in a Gibbs-Helmholtz plot (ΔGo vs. T) (Figure 5A) or a van’t Hoff plot (lnK vs. 1000/T) (Figure 5B), and reported in Table S1 of Supporting Information. Analysis by these two methods allows for easy visualization of any nonlinearity in the temperature dependence of binding for each molecular event. The overall free energy for trimeric Dpo1 binding to DNA is the sum of the free energies for the first and second binding steps, where the second “step” represents the simultaneous or cooperative binding of two additional monomers to the monomeric Dpo1-DNA complex (described in Table 2). The free energy minima for monomeric and trimeric Dpo1 binding both occur at ~61 °C. The predicted critical temperatures for TH (where ΔH = 0) and TS (where TΔS = 0) are 36 °C and 60 °C for monomeric and 41 °C and 60 °C for trimeric Dpo1, respectively. TH represents the temperature where Ka is at a maximum, and TS represents the temperature where ΔG is most favorable. This binding behavior is indicative of a temperature dependent binding enthalpy (ΔHo) with fitted heat capacity changes (ΔCop) (Equation 8) for monomer and trimeric Dpo1 binding equal to −0.43 ± 0.07 cal mol−1 K−1 and −1.02 ± 0.12 cal mol−1 K−1, respectively (Table S1 of Supporting Information). The parallel large decreases in ΔHo and TΔSo with temperature are compensatory, resulting in smaller changes in ΔGo, and are generally characteristic of sequence specific DNA binding proteins (Figure S4 A&B of Supporting Information) (29, 30, 49). It seems possible that Dpo1 and the DNA at the primer-template junction achieve a degree of structural complementarity comparable to that in sequence-specific protein-DNA interfaces, thus making a significant contribution to the large negative ΔCop. Subsequent structural specific binding of two additional molecules of Dpo1 at that site completes the trimeric Dpo1-DNA complex as identified previously by DNA footprinting (20).
Figure 5.
Thermodynamic differences of monomeric and oligomeric Dpo1 and Dpo4 binding to DNA. Gibbs-Helmholtz plots of free energy of binding (ΔGo) for A) monomeric (solid -●-, blue) or trimeric (dashed -■-, light blue) Dpo1 and C) monomeric (solid -○-, red) or dimeric (dashed -□-, pink) Dpo4 as a function of temperature. Error bars represent the standard error from multiple experiments at each point. Lines show the fits of the data to Equation 8 giving ΔCop (cal mol−1 K−1) values of for monomeric (−0.43 ± 0.07) and trimeric (−1.45 ± 0.14) Dpo1 and monomeric (−0.68 ± 0.10) and dimeric (−1.22 ± 0.15) Dpo4. van’t Hoff plots highlighting the individual binding states for B) Dpo1 or D) Dpo4. Lines show the fits to Equation 11.
Table 2.
Definition of Equilibrium Steps and Polymerase States
| Dpo1 | Dpo4 | |||
|---|---|---|---|---|
| Stepa | Kapp | ΔC°p(app) (cal mol−1 K−1) | Kapp | ΔCop(app) (cal mol−1 K−1) |
| First | K1 | −0.43 | K1 | −0.68 |
| Second | (K2)2 | −1.02 | K2 | −0.51 |
| Overallb | (K1)(K2)2 | −1.45 | (K1)(K2) | −1.22 |
As measured from fluorescence anisotropy.
Product of equilibrium constants leading to trimeric Dpo1 or dimeric Dpo4.
Dpo4 also shows a nonlinear temperature dependence of binding for both the monomer and dimer, as visualized in a Gibbs-Helmholtz plot (Figure 5C) or van’t Hoff plot (Figure 5D), and reported in Table S2 of Supporting Information. The free energy minima for monomeric and dimeric Dpo4 assemblies occur at ~49 °C, and 51 °C, respectively. The predicted critical temperatures for TH and TS are 34 °C and 49 °C for monomeric and 39 °C and 55 °C for dimeric Dpo4, respectively. The temperature at which Dpo4 binding shifts from primarily entropically driven to enthalpically driven occurs in the range of 35 °C to 40 °C and is consistent with the ITC results from Figure 2. This binding behavior is also indicative of a temperature dependent ΔHo with fitted ΔCop values (Equation 8) for monomer and dimeric Dpo4 binding of −0.68 ± 0.09 cal mol−1 K−1 and −1.22 ± 0.15 cal mol−1 K−1, respectively. Again, parallel changes in ΔH and TΔS with temperature are indicative of temperature dependent enthalpy/entropy compensation (Figure S4 C&D of Supporting Information).
Calculated ΔCp values from burial of nonpolar and polar surfaces upon Dpo4-DNA complex formation
The burial of polar and nonpolar surface areas upon binding has been used as a predictive measure relating structural details to thermodynamic parameters. Heat capacity data for the transfer of small molecule model hydrocarbons and amides from the liquid state to the aqueous solution were used to obtain an empirical relationship for the calculation of (see Equation 12) from computed values of changes in nonpolar and polar surfaces upon protein folding or protein-ligand interaction (46, 50). For the folding of many proteins and the interaction of proteins with small ligands, there has been adequate agreement between the experimentally observed ΔCp values and those predicted from burial of surface area. However, for association of macromolecules and some protein-folding reactions, there are many additional solution factors that contribute to the strongly negative observed ΔCp values. A significant source of the discrepancy between ΔCop and values is the restriction of configurational (conformational-vibrational) degrees of freedom upon association (35, 51–55).
Although the buried surface area for Dpo1 binding to DNA cannot be determined directly due to the lack of an appropriate crystal structure, 3437 Å2 of surface area are buried when a Dpo4 monomer binds to primer template DNA (44). The changes in polar (ΔAp) and nonpolar (ΔAnp) solvent accessible surface area upon Dpo4 binding to DNA are −1753 Å2 and −1683 Å2, respectively. From these values, we calculate a value of −0.29 cal mol−1 K−1 at 25 °C for monomeric Dpo4 binding to DNA from surface area contributions alone. As noted above, although the burial of nonpolar (ΔAnp) or polar (ΔAp) surface areas is often considered the most important factor contributing to ΔCop, other factors such as electrostatics, solvation, protonation, conformational strain, thermal or vibrational fluctuations, and linked equilibria can often have much larger contributions to ΔCop accounting for deviation from the experimental value (56).
Modeling temperature dependent binding populations of Dpo1 and Dpo4
Using a sequential assembly scheme with a cooperativity parameter for Dpo1 or with no cooperativity parameter for Dpo4, and the parameters from the anisotropy experiments, we are able to model the relative populations of monomer or oligomer for Dpo1 or Dpo4 bound to DNA as a function of temperature (Figure 7 and Supporting Information). Using this analysis, there is both a concentration and temperature dependent effect on the formation of monomeric or oligomeric Dpo1 or Dpo4. Below 400 nM, there is preferential binding of a monomeric Dpo1 and Dpo4 to DNA. In the range between 400 – 2000 nM, there is a mixed population of monomer and oligomeric Dpo1 or Dpo4 complexes with DNA. At concentrations greater than 2 μM, there is preferential binding of trimeric Dpo1 and dimeric Dpo4. In this analysis, it is also clear that the assemblies and populations are influenced by temperature. For Dpo1, there is an increase in affinity for both the first and second binding events up to 50 °C. Above 50 °C, there is a slight decrease in the second binding step in favor of the first. A similar trend occurs for Dpo4 with the cutoff being around 45 °C. More than 50% of the Dpo1 population exists in a trimeric state at concentrations greater than 1.5 μM while a Dpo4 concentration of 3 μM is required for 50% dimer.
Figure 7.
Concentration dependent assembly of oligomeric polymerase-DNA complexes. A) Trimeric Dpo1 assembly on DNA follows initial higher affinity binding of one molecule (K1) followed by a second step of cooperative assembly of two additional molecules (K2). B) Dimeric Dpo4 assembly on DNA that includes two sequential binding events with differing affinities (K1 and K2). Simulations of the relative populations for monomeric i) Dpo1 or Dpo4 (open symbols, representing K1) or ii) trimeric Dpo1 or dimeric Dpo4 (closed symbols, representing K2) as a function of temperature and concentrations as described in the Supporting Information. Simulations are shown for 6.8 (purple ○ or ●), 17.0 (cyan □ or ■), 27.4 (green ◇ or ◆), 38.0 (yellow △ or ▲), 48.8 (orange ▽ or ▼), and 58.9 or 59.7 (red ◺ or ◣) °C temperatures.
Dpo1 and Dpo4 Processivities Increase with Temperature and Concentration
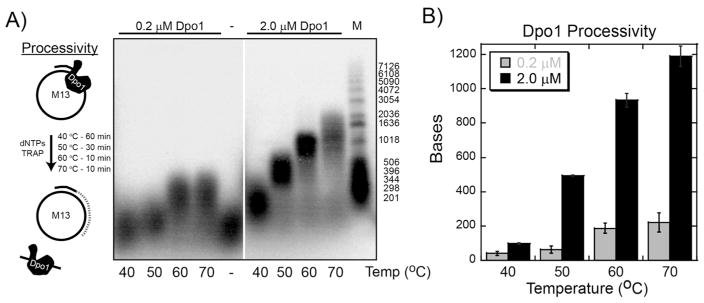
To provide a biochemical explanation for the different binding specificities for DNA polymerases across a variety of temperatures, we assayed the temperature dependent polymerization processivity for Dpo1 and Dpo4. Processivity is a measure of the stability of an enzyme substrate complex during successive catalytic steps. Processive DNA polymerases have a higher rate constant for the catalytic step of DNA synthesis (kpol) than for dissociation from the template (kdis) (57). The ratio between these kinetic parameters determines the processivity value. The processivity for Dpo4 has been measured previously over a limited concentration range of 0.5 – 200 nM representing primarily monomer, and although there is a slight increase in processivity with concentration, it was concluded that Dpo4 is essentially a distributive enzyme with processivity value of 1 to 2 nucleotides (17). Interestingly, in processivity reactions where the concentration of Dpo4 was in 20-fold excess to DNA, products of over a hundred nucleotides in length were synthesized, suggesting that polymerase interactions may modestly increase processivity. Another report of processivity when DNA template was in excess to Dpo4 (35 nM) gave a value of 16 at 37 °C (23). More strikingly, we have previously measured a large increase in processivity for Dpo1 when it is in the trimeric conformation over that of the monomeric form (20).
We have performed additional DNA polymerization experiments at various temperatures (40, 50, 60 and 70 °C) to determine if higher temperatures promote greater rates or processivities for Dpo1 due to increased binding specificity. Both kinetic and processivity experiments were performed at either 200 nM or 2.0 μM Dpo1 to represent contributions from primarily monomeric or trimeric species, respectively (Figure 8A). We chose 40 °C over room temperature experiments due to slower rates of synthesis that would limit detection. The DNA synthesis rate for trimeric Dpo1 at 40, 50, 60, and 70 °C was measured to be 36 ± 3, 209 ± 4, 447 ± 30, and 529 ± 35 bp/min, respectively, and always greater than monomeric Dpo1 rate at 36 ± 12, 76 ± 14, 366 ± 45, and 400 ± 55 bp/min, respectively, (Figure S6 of Supporting Information). Processivity experiments were initiated with the simultaneous addition of dNTPs and a high concentration of unlabeled DNA trap. Polymerase molecules that dissociate from the prebound radioactive primer-M13 template will be captured by binding to a cold DNA substrate and no longer contribute to the signal for the experiment. The concentration of trap required to be effective at all polymerase concentrations was determined empirically by titrating trap until no further increase in processivity was noted (data not shown). Reactions were incubated for different times at each temperature depending on the rate of synthesis. Dpo1 processivity at 200 nM (representing monomer) increased slightly with increasing temperature from 41 ± 12, 62 ± 14, 187 ± 45, and 220 ± 56 nucleotides at 40, 50, 60, and 70 °C, respectively, (Figure 8B). The processivity at 2 μM (representing trimer) increased more dramatically from 98 ± 2, 493 ± 5, 933 ± 39, to 1191 ± 52 nucleotides at 40, 50, 60, and 70 °C, respectively (Figure 8B). At temperatures greater than or equal to 50 °C, the processivity of the trimeric state of Dpo1 is much greater than that of the monomeric complex and reflects a change in the specificity of binding DNA consistent with the fluorescence anisotropy data presented above.
Figure 8.
Processivity of Dpo1 increases with temperature and concentration. A) Dpo1 processivity assays were performed at 40, 50, 60, and 70 °C representing monomer (0.2 μM) (left panel) or trimer (2.0 μM) (right panel) concentrations and separated on a denaturing alkaline agarose gel as described in the Materials and Methods. The inset cartoon describes the experimental protocol for processivity experiments. Longer reaction times were used for lower temperatures to compensate for slower polymerase rates. Processivity values were calculated from DNA size markers and calculated using ImageQuant software. Quantification of the processivity values (bp) comparing monomeric (200 nM, grey) or trimeric (2.0 μM, black) Dpo1 at 40, 50, 60, and 70 °C.
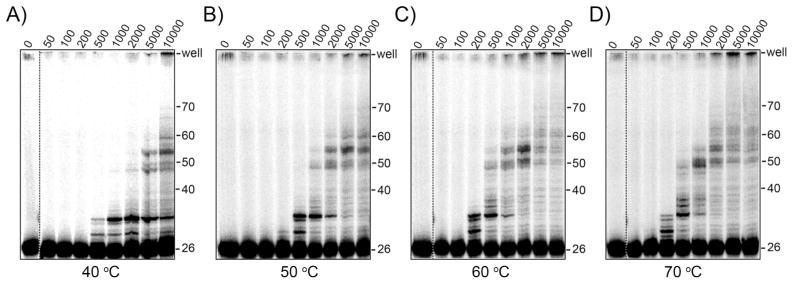
For Dpo4, processivities also increase with increasing enzyme concentration at a variety of temperatures (Figure 9) but to a lesser extent than for Dpo1. As for Dpo1, the Dpo4 processivity increases slightly at 200 nM (representing monomer) and more dramatically at 5 μM (representing dimer) with increasing temperature (Figure S7 of Supporting Information). This is not only visualized by longer products separated on the gel, but also more radioactivity seen in the wells at the higher concentrations or temperatures. Interestingly at both monomeric and dimeric Dpo4 concentrations, there is an apparent decrease in processivity when going from 60 to 70 °C consistent with decreased specificity of binding measured for Dpo4 at those temperatures using fluorescence anisotropy (Figure 5C). Similar to Dpo1, the processivity values increase when dimeric Dpo4 concentrations are used compared with monomeric Dpo4 concentrations at all temperatures.
Figure 9.
Processivity of Dpo4 increases with temperature and concentration. Dpo4 processivity assays were performed at A) 40, B) 50, C) 60, and D) 70 °C for concentrations ranging from 0.05 – 10 μM and separated on a denaturing acrylamide gel. Reactions were initiated with dNTPs and excess ssDNA trap as described in the Materials and Methods.
DISCUSSION
Using chemical crosslinking, isothermal titration calorimetry, analytical ultracentrifugation, and fluorescence anisotropy, we have been able to identify, separate, and quantify multiple binding events for DNA replication (Dpo1) and repair polymerases (Dpo4) to DNA that lead to different specificities and activities with temperature. Consistent with our previous report (20), Dpo1 forms a concentration dependent trimer at all temperatures. Unexpectedly, Dpo4 behaves similarly, forming a dimeric complex that becomes more favored at higher temperatures. For both Dpo1 and Dpo4, the affinities of monomeric and subsequent oligomeric binding generally increase as the temperature approaches the physiological range. The changes in polymerase equilibria with concentration and temperature can be clearly visualized using analytical ultracentrifugation even over a limited temperature range (10 – 30 °C). Comparison of monomeric polymerase binding to DNA shows that the differentially stronger affinity of Dpo1 than Dpo4 becomes even more exaggerated as physiological temperatures (75 °C) are approached, providing for thermodynamic selection of a DNA replication polymerase on undamaged templates. These thermodynamic results are reflected in the enzymatic behaviors, in that we measured a greater processivity for nucleotide incorporation at higher temperatures and oligomeric states for both DNA polymerases. The equilibrium microenvironment in the cell or more importantly at the replication fork will direct binding and association of a variety of DNA polymerase complexes to promote efficient DNA replication in the presence of any damage.
Evidence for Oligomeric Dpo1 and Dpo4 Complexes Bound to DNA
Identification, isolation, and assembly of the trimeric Dpo1 complex at the primer template junction have been discussed previously (20) but is now verified and quantified across a large temperature range. Similarly, crosslinking and ITC suggest that assembly of a dimeric Dpo4 is also possible. Using AU-FDS experiments, we were able to directly monitor the size distribution of polymerase DNA complexes in solution at multiple temperatures. Interestingly, for both Dpo1 and Dpo4 bound to DNA, there is a modest but reproducible shift in the sedimentation boundaries to larger coefficients with increasing temperature. Unfortunately, analytical ultracentrifugation can only be performed over a limited temperature range, 10 °C to 30 °C; as these effects may have been greater if higher temperatures could have been probed. A shift to a larger s20,w value in sedimentation velocity experiments is consistent with a change in the population of complexes towards a larger average size. The shifts in reaction boundaries occur at lower concentrations for Dpo1 than for Dpo4, consistent with the anisotropy result that Dpo1 has a higher affinity for DNA than Dpo4 at identical temperatures. Distinct s20,w populations are more easily seen for Dpo1, due to large differences in the molecular weight between the complexes of monomer and trimer with DNA. For Dpo4, the size difference between monomeric and dimeric bound states is much less, causing a general broadening of the s20,w distribution and discrete s20,w weight average values for each state are not observed. The most obvious temperature dependent reaction boundary shifts occur at concentrations equal to the dissociation constant for polymerase binding to DNA (i.e. 100 nM Dpo4 at 30 °C in Figure S2C of Supporting Information). Shifts in the reaction boundaries of Dpo1 bound to DNA with increasing temperature are more subtle, but reproducible, in this experimental range; these shifts are characterized by better resolution between monomeric and trimeric Dpo1 at 30 °C (Figure 3C). Although only qualitatively, the individual sedimentation boundaries correlate well with the Kd1 and Kd2 binding affinities (K1 and K2 in Figure 7) for Dpo1 and Dpo4 to DNA measured by fluorescence anisotropy. Sedimentation equilibrium experiments would be useful in quantifying the actual populations for either Dpo1 or Dpo4 alone or bound to DNA, but unfortunately resulted in uninterpretable spectra, probably due to some precipitation or aggregation during the long times required to attain sedimentation equilibrium. No loss in spectral signal was detected in the analytical velocity experiments suggesting that aggregation and precipitation is not an issue for shorter time scales.
Although there are a number of biochemical, kinetic and structural papers involving the mechanism of action for Dpo4 (17, 18, 23, 24, 44, 58–62), none of them suggest that a dimeric DNA polymerase complex is the active species. However, we have confirmed Dpo4 binding to DNA as a dimer using chemical crosslinking, the stoichiometry values from ITC, and analytical ultracentrifugation. The apparent dissociation constants measured by ITC at or are much larger than expected based on AUC, fluorescence anisotropy and EMSA results previously published (63–65) and most likely represent contributions of equilibria from monomer and dimer Dpo4 binding to DNA. The analytical ultracentrifugation results show that binding begins at a concentration of 100 nM and then proceeds in a concentration dependent manner towards dimer above 500 nM. According to the fits of the fluorescence anisotropy experiments, dimer assembly persists across a range of temperatures. From a variety of Dpo4:DNA X-ray structures, the site size of Dpo4 on DNA consists of roughly 10 bases of dsDNA and 4 bases of ssDNA straddling the primer template junction (18). The DNA hairpin primer-template used in these studies has a 12 base pair duplex and a 10 base single strand region, thus restricting binding site size to a single DNA polymerase. The location of the protein crosslink can be pinpointed because there is only a single native cysteine residue (C31) in Dpo4 but does not exclude other dimeric Dpo4 structures. Unlike what we observed for Dpo1 (20), the presence of DNA did not significantly affect the degree of crosslinking, suggesting that at the concentrations used for this experiment, Dpo4 can form a dimer in the absence of DNA.
Within the RCSB Protein Data Bank (www.rcsb.org), there exist roughly 100 structures of Dpo4 both without DNA and bound to various types of DNA templates (damaged and undamaged). Roughly, one-third of these structures have multiple Dpo4 molecules interacting in the crystal unit in various conformations. Many of these multimeric structures show Dpo4 in a conformation that would allow the cysteine residues in the interface to be in close enough proximity for crosslinking to occur (66–68). The rest of the oligomeric Dpo4 structures are in a variety of alternative dimeric or tetrameric complexes. Additionally, a number of structures for the analogous Dbh polymerase from Sulfolobus acidocaldarius also show oligomeric complexes in the crystal unit with the homologous cysteine residue in close enough proximity for crosslinking (69, 70). Although we cannot be certain of the exact conformation, we are able to detect and verify a previously unrecognized dimeric Dpo4 complex across all temperature ranges that is consistent with our thermodynamic binding data. Moreover, the variety of dimeric and tetrameric states of Dpo4 seen by X-ray crystallography may suggest that the binding equilibria are even more complex than we include in our model (Figure 7B).
Thermodynamic differences in binding oligomeric replication and repair polymerases to primer-template DNA
In fluorescence anisotropy experiments, optimum fits to binding isotherms for the titration of Dpo1 or Dpo4 with DNA are obtained using a model for a sequential assembly path involving two binding events. Information from crosslinking, EMSA, gel filtration, AUC, and ITC (20) experiments about the initial (monomeric) and final (trimeric for Dpo1 and dimeric for Dpo4) forms of polymerase-DNA complexes was essential for differentiating between single and multiple binding events in the anisotropy experiments. For Dpo1 and Dpo4, there is an initial higher affinity binding of one polymerase molecule to DNA. The binding of the second and third molecule of Dpo1 to complete the trimer is proposed to occur cooperatively (20); however since a dimeric Dpo1-DNA complex cannot be resolved, our data are insufficient to separate these secondary binding events. Formation of a Dpo4 dimer proceeds through two sequential binding events. For both Dpo1 and Dpo4, binding of additional polymerase molecule(s) to the first is inferred primarily from the limited DNA template size and the direct contacts found using chemical crosslinking (20). Previous reports on binding affinity for Dpo1 (71) and Dpo4 (65) are consistent with our values for monomeric assembly at room temperature but those studies did not test higher concentrations required for multimeric assemblies.
A comparison of the DNA binding affinities of monomeric Dpo1 and Dpo4 shows that binding affinity is only slightly more favorable for monomeric Dpo1 across all temperature ranges but becomes more selective at physiological temperatures (Figure 6 and Figure S5 of Supporting Information). The free energy change for binding the first molecule of Dpo1 decreases steadily with temperature up to at least ~65 °C, where binding is preferred by about −0.8 kcal mol−1 over Dpo4. Dpo4, on the other hand, has a free energy binding minimum around 50 °C, disfavoring binding to undamaged DNA in the presence of Dpo1. Dpo4 is smaller, known to have a more open active site than typical B-family polymerases, exists in two distinct conformations, and has subtle repositioning of active site residues upon binding (18, 44, 72, 73). The little finger domain and associated linker in particular seem to be most important for stable binding to DNA. On the other hand, the binding affinity of Dpo1 to DNA is more favorable at higher temperature, consistent with formation of a tight closed conformation on DNA, resulting in greater DNA stabilizing ability/annealing noted previously (74).
There is a larger differential in the free energies of binding of the oligomeric forms of the polymerases (trimeric Dpo1 or dimeric Dpo4) (Figure 6 and Figure S5 of Supporting Information) than for the monomeric forms. The ΔG° for formation of a trimeric Dpo1 complex is much more favorable than that for formation of a dimeric Dpo4 complex. The difference in binding energies becomes even more exaggerated at higher temperatures, thus increasingly favoring the trimeric Dpo1 complex at physiological temperatures. At 64 °C, binding of trimeric Dpo1 is enormously favored (by about −9.3 kcal/mol) over dimeric Dpo4. The slight preference for binding primer-templates by the Dpo1 monomer (over that of the Dpo4 monomer) will lead to trimeric Dpo1 complex formation, thus selecting against Dpo4 binding.
Formation of both monomeric and oligomeric polymerase-DNA complexes produces large negative Δ COP values
Amongst the thermodynamic parameters (ΔG, ΔH, ΔS, ΔCP, and volume) ΔCp is one of the least well known, but potentially the most informative with respect to extracting molecular information about specificity. Because ΔCp was first analyzed for protein folding, the working hypothesis was that the net negative ΔCp reflected primarily the burial of nonpolar surface area (46, 50). However, a number of studies on site-specific protein-DNA complexes have shown large deficits between the values predicted from surface burial and experimental ΔCop values (29, 30, 35, 49, 52–54, 56). In addition to desolvation of nonpolar surface upon binding, other factors such as restriction of conformational-vibrational motions of the macromolecules, interfacial waters, and linkage of other binding equilibria (protonation, cation and anion binding, and conformational changes) contribute to the protein-DNA binding reaction and can potentially account for the deficit between and ΔCop (29, 30, 75–82). Although a strongly negative ΔCoP value has generally been considered a key signature of sequence-specific DNA-protein interactions (29), large negative values of ΔCoP have also been observed for the formation of interfaces with high structural complementarity between DNA polymerases and their primer-template substrates (31, 32, 83, 84).
In the absence of appropriate monomeric or oligomeric polymerase-DNA structures, contributions of surface area burial cannot be directly assessed. The only appropriate data set is for a monomeric Dpo4-DNA structure, which underestimates the contributions of buried surface area ( ) to the experimental value. The ΔCoP values are similar for monomeric Dpo1 (−0.43 cal mol−1 K−1) and Dpo4 (−0.68 cal mol−1 K−1) but significantly more negative than the value suggesting that other factors in addition to surface area burial contribute to the experimental ΔCop values.
Because we have been able to monitor the sequential steps in polymerase binding, we have also found that the ΔCop values for assembly of trimeric Dpo1and dimeric Dpo4 complexes are strongly negative, suggesting that structure-specific binding is occurring during formation of these oligomeric DNA complexes as well. The sign and magnitude of the ΔCop values for formation of Dpo1 and Dpo4 oligomers are consistent with other specific dimerization (85) or binary protein binding events (86–88). No structural information is available for an oligomeric Dpo1 complex, nor can we be certain of the molecular arrangement of a dimeric Dpo4 complex, which makes calculation of buried surface area difficult. Nevertheless, exclusion of water molecules, favorable surface interactions between the polymerase molecules, and restriction of configurational freedom within the oligomeric complex are consistent with the magnitudes for the oligomeric polymerase-DNA ΔCop values. Therefore, many of the factors discussed above may contribute to the strongly negative ΔCoP values observed for the formation of trimeric Dpo1 and dimeric Dpo4 complexes with primer-template DNA substrates.
Oligomeric DNA Polymerases Have Increased Activities and Processivities
Generally, processivity is thought to be a temperature independent parameter although slight decreases in processivity with increasing temperature have been noted for the telomerase enzyme (89). DNA polymerases alone are fairly distributive enzymes unless accompanied by their respective circular clamp proteins which can increase processivity from less than 20 to greater than 10,000 bases (90). Previously, we found that the processivity of Dpo1 was dependent on concentration, such that that trimeric complex had much greater processivity (~1000 bases) than the monomer at 60 °C (20). We have now shown that the processivity for both Dpo1 and Dpo4 increases with temperature. An increase in processivity with temperature was noted for both monomeric and oligomeric complexes of Dpo1 and Dpo4 although the effect was more dramatic for the oligomeric states. Trimeric Dpo1 was 4–5 fold more processive than monomeric Dpo1; dimeric Dpo4 was significantly more processive than monomeric Dpo4; but the processivity of trimeric Dpo1 was more than 15-fold greater than that of dimeric Dpo4 at higher temperatures. Even though Dpo4 is generally considered to be a distributive enzyme, the increased affinities for binding DNA noted with increasing temperatures and concentrations also increase the processivity. We reason that for Dpo1 and Dpo4, a more tightly bound monomeric or oligomeric complex promotes greater processivities at higher temperatures. For Dpo1, the affinity for DNA generally increases with temperature, and formation of a trimeric Dpo1 reduces the off-rate of the complex from DNA over monomeric Dpo1 (20) explaining the larger processivity values. The combination of higher intrinsic processivity for monomeric Dpo1 compared with Dpo4, as well as increased binding affinity at higher temperatures of the Dpo1 trimer, contribute to the enzymatic activity resulting in high trimeric Dpo1 processivity. In fact, phi29 is the only other characterized DNA polymerase with a greater processivity value and acts analogously by topologically encircling the DNA template (91, 92).
Increased binding affinity of Dpo4 to DNA also correlates well with increasing processivity up to 60 °C. At 70 °C, the processivity decreases slightly, consistent with the measured affinity values. Previously, when high concentrations of Dpo4 were used, an increase in the length of product synthesized was observed suggesting that either faster repeated binding was occurring in the absence of trap or cooperation between molecules at higher concentrations afforded greater processivity (73). The authors implicated the little finger domain of Dpo4 in maintaining moderate processivity by creating a closed more stable enzyme complex on DNA. The conformational change of the little finger domain is considered to be the rate limiting step and occurs both before and after chemistry (23, 93). Kinetic experiments have shown that of the 7 steps within the catalytic cycle for a single nucleotide incorporation event, the conformational change step that precedes chemistry is most affected by temperature increasing 20-fold from 37 to 56 °C (59). Moreover, the little finger domains are in close proximity with one another in a variety of dimeric and tetrameric Dpo4 crystal structures including our crosslinking model in Figure 1B, suggesting that dimerization may stabilize a closed complex, decrease the off-rate, and increase the rate limiting conformational change step to positively affect processivity.
Clearly, a tightly bound trimeric Dpo1 complex will increase the speed and processivity of polymerization and may be utilized in various genomic maintenance applications. At 75 °C, binding of Dpo4 will be disfavored on undamaged primer-templates where Dpo1 is directing synthesis. Selection and increased processivity will also be provided through interactions with the processivity clamp, SsoPCNA123, but the affinities of Dpo1 for PCNA2 (94) and Dpo4 for PCNA1 (95) are very similar (Trakselis, unpublished). Switching from Dpo1 to Dpo4 will depend on a change in the thermodynamics of binding either due to polymerase stalling, repeated shuttling between polymerase and exonuclease sites, detection of DNA damage, or a change in the local concentrations. In those cases, binding of Dpo4 would become preferred. It has been recently estimated that the concentration of Dpo1 is at least an order of magnitude greater than that of Dpo4 in the cell (63) suggesting that Dpo1 will be preferentially bound and will have a significant population of trimer at the replication fork. Interestingly, mRNA levels of Dpo1 decrease when cells are exposed to DNA damage in favor of Dpo4 and another B-family DNA polymerase (Dpo2) (96, 97), suggesting that equilibrium changes will direct appropriate binding of the required DNA polymerase.
Although the oligomeric state of Dpo1 modulates both the speed and processivity of replication, the biological role of a dimeric Dpo4 remains elusive. Due to only slight increases in biochemical activity, we would predict that a dimeric Dpo4 would not be essential for cellular catalysis, but rather in either increasing the concentration of DNA polymerases at sites of DNA damage or stabilizing the closed conformational state promoting catalysis. Accurate and efficient DNA replication at high temperatures requires minimal differences in the thermodynamics of DNA polymerase binding to DNA for easy exchange of enzymes for uninterrupted synthesis. This thermodynamic compensation will be affected by small changes in the cellular equilibria that direct formation of higher order protein complexes that promote a variety of genomic maintenance activities. The detection of multimeric polymerase complexes for both Dpo1 and Dpo4 suggests a possible mechanism for exchange, whereby direct interactions between polymerases maintain high local concentrations at the replication fork that can thermodynamically switch binding modes when required.
Supplementary Material
Acknowledgments
Funding: This work was supported by startup funds from the University of Pittsburgh, Department of Chemistry (M.A.T.), a Research Scholar Grant (RSG-11-049-01-DMC) to. M.A.T. from the American Cancer Society, National Institutes of Health MERIT award (5R37-GM29207) to L.J.-J. Funding for the AUC was provided by the National Science Foundation (NSF, BIR-9876582) and the National Institutes of Health (NIH, 1R01GM6283601) to T.M.L.
We thank Roger Woodgate for providing the pET22b-Dpo4 expression plasmid. We also thank Reza Salari and Lillian Chong for help in calculating buried surface areas for Dpo4 and DNA.
Abbreviations
- ITC
Isothermal titration calorimetry
- ΔCop
change in heat capacities
- ΔCpASA
surface area calculated heat capacity
- ΔAp
change in polar surface area
- ΔAnp
change in nonpolar surface area
- CPS
counts per second
- Kd
dissociation constant
- Kapp
apparent association constant
- ΔGo
free energy
- ΔHo
enthalpy
- ΔSo
entropy
- n
stoichiometry
- AUC
analytical ultracentrifugation
- AU-FDS
analytical ultracentrifugation fluorescence-detected sedimentation
- Dt
diffusion coefficient
- s
sedimentation coefficient
- EMSA
electrophoretic mobility shift assay
- kpol
rate constant of synthesis for the next catalytic step
- kdis
rate constant of dissociation from the template
- nt
nucleotide
Footnotes
SUPPORTING INFORMATION AVAILABLE
Simulation data for concentration and temperature dependent assembly of Dpo1 and Dpo4 on DNA. Tables S1 and S2 contain the thermodynamic parameters for Dpo1 and Dpo4 binding to DNA, respectively. Figure S1 (AUC data of Dpo1 and Dpo4 alone), Figure S2 (AU-FDS temperature dependent data of Dpo1 and Dpo4 bound to DNA), Figure S3 (examples of anisotropy fits), Figure S4 (fitted thermodynamic parameters for Dpo1 and Dpo4 binding to DNA), Figure S5 (Thermodynamic differences between Dpo1 and Dpo4 binding to DNA), Figure S6 (Dpo1 polymerization rate at different temperatures), and Figure S7 (Dpo4 processivity at different temperatures) are available. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Burgers PM, Koonin EV, Bruford E, Blanco L, Burtis KC, Christman MF, Copeland WC, Friedberg EC, Hanaoka F, Hinkle DC, Lawrence CW, Nakanishi M, Ohmori H, Prakash L, Prakash S, Reynaud CA, Sugino A, Todo T, Wang Z, Weill JC, Woodgate R. Eukaryotic DNA polymerases: Proposal for a revised nomenclature. J Biol Chem. 2001;276:43487–43490. doi: 10.1074/jbc.R100056200. [DOI] [PubMed] [Google Scholar]
- 2.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishida H, Mayanagi K, Kiyonari S, Sato Y, Oyama T, Ishino Y, Morikawa K. Structural determinant for switching between the polymerase and exonuclease modes in the PCNA-replicative DNA polymerase complex. Proc Natl Acad Sci U S A. 2009;106:20693–20698. doi: 10.1073/pnas.0907780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purohit V, Grindley ND, Joyce CM. Use of 2-aminopurine fluorescence to examine conformational changes during nucleotide incorporation by DNA polymerase I (Klenow fragment) Biochemistry. 2003;42:10200–10211. doi: 10.1021/bi0341206. [DOI] [PubMed] [Google Scholar]
- 5.Ishmael FT, Trakselis MA, Benkovic SJ. Protein-protein interactions in the bacteriophage T4 replisome. The leading strand holoenzyme is physically linked to the lagging strand holoenzyme and the primosome. J Biol Chem. 2003;278:3145–3152. doi: 10.1074/jbc.M209858200. [DOI] [PubMed] [Google Scholar]
- 6.Jezewska MJ, Rajendran S, Bujalowski W. Transition between different binding modes in rat DNA polymerase beta- ssDNA complexes. J Mol Biol. 1998;284:1113–1131. doi: 10.1006/jmbi.1998.2252. [DOI] [PubMed] [Google Scholar]
- 7.Tang KH, Niebuhr M, Aulabaugh A, Tsai MD. Solution structures of 2 : 1 and 1 : 1 DNA polymerase-DNA complexes probed by ultracentrifugation and small-angle X-ray scattering. Nucleic Acids Res. 2008;36:849–860. doi: 10.1093/nar/gkm1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jezewska MJ, Bujalowski PJ, Bujalowski W. Interactions of the DNA polymerase X from African swine fever virus with gapped DNA substrates. Quantitative analysis of functional structures of the formed complexes. Biochemistry. 2007;46:12909–12924. doi: 10.1021/bi700677j. [DOI] [PubMed] [Google Scholar]
- 9.Tang KH, Tsai MD. Structure and function of 2:1 DNA polymerase. DNA complexes. J Cell Physiol. 2008;216:315–320. doi: 10.1002/jcp.21458. [DOI] [PubMed] [Google Scholar]
- 10.McInerney P, Johnson A, Katz F, O'Donnell M. Characterization of a triple DNA polymerase replisome. Mol Cell. 2007;27:527–538. doi: 10.1016/j.molcel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 11.Yang J, Zhuang Z, Roccasecca RM, Trakselis MA, Benkovic SJ. The dynamic processivity of the T4 DNA polymerase during replication. Proc Natl Acad Sci U S A. 2004;101:8289–8294. doi: 10.1073/pnas.0402625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loparo JJ, Kulczyk AW, Richardson CC, van Oijen AM. Simultaneous single-molecule measurements of phage T7 replisome composition and function reveal the mechanism of polymerase exchange. Proc Natl Acad Sci U S A. 2011;108:3584–3589. doi: 10.1073/pnas.1018824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koonin EV, Mushegian AR, Galperin MY, Walker DR. Comparison of archaeal and bacterial genomes: Computer analysis of protein sequences predicts novel functions and suggests a chimeric origin for the archaea. Mol Microbiol. 1997;25:619–637. doi: 10.1046/j.1365-2958.1997.4821861.x. [DOI] [PubMed] [Google Scholar]
- 14.Barry ER, Bell SD. DNA replication in the archaea. Microbiol Mol Biol Rev. 2006;70:876–887. doi: 10.1128/MMBR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tahirov TH, Makarova KS, Rogozin IB, Pavlov YI, Koonin EV. Evolution of DNA polymerases: An inactivated polymerase-exonuclease module in Pol epsilon and a chimeric origin of eukaryotic polymerases from two classes of archaeal ancestors. Biol Direct. 2009;4:11. doi: 10.1186/1745-6150-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savino C, Federici L, Johnson KA, Vallone B, Nastopoulos V, Rossi M, Pisani FM, Tsernoglou D. Insights into DNA replication: The crystal structure of DNA polymerase B1 from the archaeon Sulfolobus solfataricus. Structure. 2004;12:2001–2008. doi: 10.1016/j.str.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Boudsocq F, Iwai S, Hanaoka F, Woodgate R. Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): An archaeal DinB-like DNA polymerase with lesion-bypass properties akin to eukaryotic poleta. Nucleic Acids Res. 2001;29:4607–4616. doi: 10.1093/nar/29.22.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 19.Kokoska RJ, Bebenek K, Boudsocq F, Woodgate R, Kunkel TA. Low fidelity DNA synthesis by a Y-family DNA polymerase due to misalignment in the active site. J Biol Chem. 2002;277:19633–19638. doi: 10.1074/jbc.M202021200. [DOI] [PubMed] [Google Scholar]
- 20.Mikheikin AL, Lin HK, Mehta P, Jen-Jacobson L, Trakselis MA. A trimeric DNA polymerase complex increases the native replication processivity. Nucleic Acids Res. 2009;37:7194–7205. doi: 10.1093/nar/gkp767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown JA, Suo Z. Elucidating the kinetic mechanism of DNA polymerization catalyzed by Sulfolobus solfataricus P2 DNA polymerase B1. Biochemistry. 2009;48:7502–7511. doi: 10.1021/bi9005336. [DOI] [PubMed] [Google Scholar]
- 22.Fiala KA, Hypes CD, Suo Z. Mechanism of abasic lesion bypass catalyzed by a Y-family DNA polymerase. J Biol Chem. 2007;282:8188–8198. doi: 10.1074/jbc.M610718200. [DOI] [PubMed] [Google Scholar]
- 23.Fiala KA, Suo Z. Mechanism of DNA polymerization catalyzed by Sulfolobus solfataricus P2 DNA polymerase IV. Biochemistry. 2004;43:2116–2125. doi: 10.1021/bi035746z. [DOI] [PubMed] [Google Scholar]
- 24.Fiala KA, Suo Z. Pre-steady-state kinetic studies of the fidelity of Sulfolobus solfataricus P2 DNA polymerase IV. Biochemistry. 2004;43:2106–2115. doi: 10.1021/bi0357457. [DOI] [PubMed] [Google Scholar]
- 25.Zang H, Irimia A, Choi JY, Angel KC, Loukachevitch LV, Egli M, Guengerich FP. Efficient and high fidelity incorporation of dCTP opposite 7,8-dihydro-8-oxodeoxyguanosine by Sulfolobus solfataricus DNA polymerase Dpo4. J Biol Chem. 2006;281:2358–2372. doi: 10.1074/jbc.M510889200. [DOI] [PubMed] [Google Scholar]
- 26.Zang H, Goodenough AK, Choi JY, Irimia A, Loukachevitch LV, Kozekov ID, Angel KC, Rizzo CJ, Egli M, Guengerich FP. DNA adduct bypass polymerization by Sulfolobus solfataricus DNA polymerase Dpo4: Analysis and crystal structures of multiple base pair substitution and frameshift products with the adduct 1,N2-ethenoguanine. J Biol Chem. 2005;280:29750–29764. doi: 10.1074/jbc.M504756200. [DOI] [PubMed] [Google Scholar]
- 27.Sakofsky CJ, Foster PL, Grogan DW. Roles of the Y-family DNA polymerase Dbh in accurate replication of the Sulfolobus genome at high temperature. DNA Repair (Amst) 2012;11:391–400. doi: 10.1016/j.dnarep.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Felice M, Medagli B, Esposito L, De Falco M, Pucci B, Rossi M, Gruz P, Nohmi T, Pisani FM. Biochemical evidence of a physical interaction between Sulfolobus solfataricus B-family and Y-family DNA polymerases. Extremophiles. 2007;11:277–282. doi: 10.1007/s00792-006-0038-x. [DOI] [PubMed] [Google Scholar]
- 29.Jen-Jacobson L, Engler LE, Jacobson LA. Structural and thermodynamic strategies for site-specific DNA binding proteins. Structure. 2000;8:1015–1023. doi: 10.1016/s0969-2126(00)00501-3. [DOI] [PubMed] [Google Scholar]
- 30.Jen-Jacobson L, Engler LE, Ames JT, Kurpiewski MR, Grigorescu A. Thermodynamic parameters of specific and nonspecific protein-DNA binding. Supramolecular Chemistry. 2000;12:143–160. [Google Scholar]
- 31.Datta K, LiCata VJ. Thermodynamics of the binding of Thermus aquaticus DNA polymerase to primed-template DNA. Nucleic Acids Res. 2003;31:5590–5597. doi: 10.1093/nar/gkg774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Datta K, Wowor AJ, Richard AJ, LiCata VJ. Temperature dependence and thermodynamics of Klenow polymerase binding to primed-template DNA. Biophys J. 2006;90:1739–1751. doi: 10.1529/biophysj.105.071837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeda Y, Ross PD, Mudd CP. Thermodynamics of Cro protein-DNA interactions. Proc Natl Acad Sci USA. 1992;89:8180–8184. doi: 10.1073/pnas.89.17.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lundback T, Hansson H, Knapp S, Ladenstein R, Hard T. Thermodynamic characterization of non-sequence-specific DNA-binding by the Sso7d protein from Sulfolobus solfataricus. J Mol Biol. 1998;276:775–786. doi: 10.1006/jmbi.1997.1558. [DOI] [PubMed] [Google Scholar]
- 35.Ladbury JE, Wright JG, Sturtevant JM, Sigler PB. A thermodynamic study of the trp repressor-operator interaction. J Mol Biol. 1994;238:669–681. doi: 10.1006/jmbi.1994.1328. [DOI] [PubMed] [Google Scholar]
- 36.Eom SH, Wang J, Steitz TA. Structure of Taq polymerase with DNA at the polymerase active site. Nature. 1996;382:278–281. doi: 10.1038/382278a0. [DOI] [PubMed] [Google Scholar]
- 37.Beese LS, Derbyshire V, Steitz TA. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993;260:352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- 38.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 39.Pierce MM, Raman CS, Nall BT. Isothermal titration calorimetry of protein-protein interactions. Methods. 1999;19:213–221. doi: 10.1006/meth.1999.0852. [DOI] [PubMed] [Google Scholar]
- 40.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laue TM, Saha BD, Ridgeway TM, Pellettier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, editor. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry; Cambridge, UK: 1992. pp. 90–125. [Google Scholar]
- 42.Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- 43.Marras SA. Selection of fluorophore and quencher pairs for fluorescent nucleic acid hybridization probes. Methods Mol Biol. 2006;335:3–16. doi: 10.1385/1-59745-069-3:3. [DOI] [PubMed] [Google Scholar]
- 44.Wong JH, Fiala KA, Suo Z, Ling H. Snapshots of a Y-family DNA polymerase in replication: Substrate-induced conformational transitions and implications for fidelity of Dpo4. J Mol Biol. 2008;379:317–330. doi: 10.1016/j.jmb.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 45.Sanner MF, Olson AJ, Spehner JC. Reduced surface: An efficient way to compute molecular surfaces. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 46.Spolar RS, Livingstone JR, Record MT. Use of liquid hydrocarbon and amide transfer data to estimate contributions to thermodynamic functions of protein folding from the removal of nonpolar and polar surface from water. Biochemistry. 1992;31:3947–3955. doi: 10.1021/bi00131a009. [DOI] [PubMed] [Google Scholar]
- 47.Zhang H, Eoff RL, Kozekov ID, Rizzo CJ, Egli M, Guengerich FP. Structure-function relationships in miscoding by Sulfolobus solfataricus DNA polymerase Dpo4: Guanine N2,N2-dimethyl substitution produces inactive and miscoding polymerase complexes. J Biol Chem. 2009;284:17687–17699. doi: 10.1074/jbc.M109014274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kingsbury JS, Laue TM. Fluorescence-detected sedimentation in dilute and highly concentrated solutions. Methods Enzymol. 2011;492:283–304. doi: 10.1016/B978-0-12-381268-1.00021-5. [DOI] [PubMed] [Google Scholar]
- 49.Ha JH, Spolar RS, Record MT., Jr Role of the hydrophobic effect in stability of site-specific protein-DNA complexes. J Mol Biol. 1989;209:801–816. doi: 10.1016/0022-2836(89)90608-6. [DOI] [PubMed] [Google Scholar]
- 50.Spolar RS, Record MT., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 51.Lazaridis T, Karplus M. Heat capacity and compactness of denatured proteins. Biophys Chem. 1999;78:207–217. doi: 10.1016/s0301-4622(99)00022-8. [DOI] [PubMed] [Google Scholar]
- 52.Petri V, Hsieh M, Brenowitz M. Thermodynamic and kinetic characterization of the binding of the TATA binding protein to the adenovirus E4 promoter. Biochemistry. 1995;34:9977–9984. doi: 10.1021/bi00031a020. [DOI] [PubMed] [Google Scholar]
- 53.Berger C, Jelesarov I, Bosshard HR. Coupled folding and site-specific binding of the GCN4-bZIP transcription factor to the AP-1 and ATF/CREB DNA sites studied by microcalorimetry. Biochemistry. 1996;35:14984–14991. doi: 10.1021/bi961312a. [DOI] [PubMed] [Google Scholar]
- 54.Lundback T, Cairns C, Gustafsson JA, Carlstedt-Duke J, Hard T. Thermodynamics of the glucocorticoid receptor-DNA interaction: binding of wild-type GR DBD to different response elements. Biochemistry. 1993;32:5074–5082. doi: 10.1021/bi00070a015. [DOI] [PubMed] [Google Scholar]
- 55.Morton CJ, Ladbury JE. Water-mediated protein-DNA interactions: The relationship of thermodynamics to structural detail. Protein Science. 1996;5:2115–2118. doi: 10.1002/pro.5560051018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jen-Jacobson L, Jacobson LA. Protein-Nucleic Acid Interactions: Structural Biology. The Royal Society of Chemistry; 2008. Chapter 2 Role of Water and Effects of Small Ions in Site-specific Protein-DNA Interactions; pp. 13–46. [Google Scholar]
- 57.Fairfield FR, Newport JW, Dolejsi MK, von Hippel PH. On the processivity of DNA replication. J Biomol Struct Dyn. 1983;1:715–727. doi: 10.1080/07391102.1983.10507477. [DOI] [PubMed] [Google Scholar]
- 58.Eoff RL, Sanchez-Ponce R, Guengerich FP. Conformational changes during nucleotide selection by Sulfolobus solfataricus DNA polymerase Dpo4. J Biol Chem. 2009;284:21090–21099. doi: 10.1074/jbc.M109.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fiala KA, Sherrer SM, Brown JA, Suo Z. Mechanistic consequences of temperature on DNA polymerization catalyzed by a Y-family DNA polymerase. Nucleic Acids Res. 2008;36:1990–2001. doi: 10.1093/nar/gkn004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fiala KA, Suo Z. Sloppy bypass of an abasic lesion catalyzed by a Y-family DNA polymerase. J Biol Chem. 2007;282:8199–8206. doi: 10.1074/jbc.M610719200. [DOI] [PubMed] [Google Scholar]
- 61.Vaisman A, Ling H, Woodgate R, Yang W. Fidelity of Dpo4: Effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 2005;24:2957–2967. doi: 10.1038/sj.emboj.7600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ling H, Boudsocq F, Woodgate R, Yang W. Snapshots of replication through an abasic lesion; structural basis for base substitutions and frameshifts. Mol Cell. 2004;13:751–762. doi: 10.1016/s1097-2765(04)00101-7. [DOI] [PubMed] [Google Scholar]
- 63.Choi JY, Eoff RL, Pence MG, Wang J, Martin MV, Kim EJ, Folkmann LM, Guengerich FP. Roles of the four DNA polymerases of the crenarchaeon Sulfolobus solfataricus and accessory proteins in DNA replication. J Biol Chem. 2011;286:31180–31193. doi: 10.1074/jbc.M111.258038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sherrer SM, Brown JA, Pack LR, Jasti VP, Fowler JD, Basu AK, Suo Z. Mechanistic studies of the bypass of a bulky single–base lesion catalyzed by a Y–family DNA polymerase. J Biol Chem. 2009;284:6379–6388. doi: 10.1074/jbc.M808161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fiala KA, Brown JA, Ling H, Kshetry AK, Zhang J, Taylor JS, Yang W, Suo Z. Mechanism of template-independent nucleotide incorporation catalyzed by a template-dependent DNA polymerase. J Mol Biol. 2007;365:590–602. doi: 10.1016/j.jmb.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eoff RL, Stafford JB, Szekely J, Rizzo CJ, Egli M, Guengerich FP, Marnett LJ. Structural and functional analysis of Sulfolobus solfataricus Y-family DNA polymerase Dpo4-catalyzed bypass of the malondialdehyde-deoxyguanosine adduct. Biochemistry. 2009;48:7079–7088. doi: 10.1021/bi9003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Irimia A, Eoff RL, Pallan PS, Guengerich FP, Egli M. Structure and activity of Y-class DNA polymerase Dpo4 from Sulfolobus solfataricus with templates containing the hydrophobic thymine analog 2,4-difluorotoluene. J Biol Chem. 2007;282:36421–36433. doi: 10.1074/jbc.M707267200. [DOI] [PubMed] [Google Scholar]
- 68.Silvian LF, Toth EA, Pham P, Goodman MF, Ellenberger T. Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus. Nat Struct Biol. 2001;8:984–989. doi: 10.1038/nsb1101-984. [DOI] [PubMed] [Google Scholar]
- 69.Wilson RC, Pata JD. Structural insights into the generation of single-base deletions by the Y family DNA polymerase dbh. Mol Cell. 2008;29:767–779. doi: 10.1016/j.molcel.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 70.Pata JD. Structural diversity of the Y-family DNA polymerases. Biochim Biophys Acta. 2010;1804:1124–1135. doi: 10.1016/j.bbapap.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gruz P, Shimizu M, Pisani FM, De FM, Kanke Y, Nohmi T. Processing of DNA lesions by archaeal DNA polymerases from Sulfolobus solfataricus. Nucleic Acids Res. 2003;31:4024–4030. doi: 10.1093/nar/gkg447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu C, Maxwell BA, Brown JA, Zhang L, Suo Z. Global conformational dynamics of a Y-family DNA polymerase during catalysis. PLoS Biol. 2009;7:e1000225. doi: 10.1371/journal.pbio.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boudsocq F, Kokoska RJ, Plosky BS, Vaisman A, Ling H, Kunkel TA, Yang W, Woodgate R. Investigating the role of the little finger domain of Y-family DNA polymerases in low fidelity synthesis and translesion replication. J Biol Chem. 2004;279:32932–32940. doi: 10.1074/jbc.M405249200. [DOI] [PubMed] [Google Scholar]
- 74.Zuo Z, Lin HK, Trakselis MA. Strand annealing and terminal transferase activities of a B-family DNA polymerase. Biochemistry. 2011;50:5379–5390. doi: 10.1021/bi200421g. [DOI] [PubMed] [Google Scholar]
- 75.Sturtevant JM. Heat capacity and entropy changes in processes involving proteins. Proc Natl Acad Sci U S A. 1977;74:2236–2240. doi: 10.1073/pnas.74.6.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eftink MR, Anusiem AC, Biltonen RL. Enthalpy-entropy compensation and heat capacity changes for protein-ligand interactions: General thermodynamic models and data for the binding of nucleotides to ribonuclease A. Biochemistry. 1983;22:3884–3896. doi: 10.1021/bi00285a025. [DOI] [PubMed] [Google Scholar]
- 77.Peters WB, Edmondson SP, Shriver JW. Effect of mutation of the Sac7d intercalating residues on the temperature dependence of DNA distortion and binding thermodynamics. Biochemistry. 2005;44:4794–4804. doi: 10.1021/bi047382w. [DOI] [PubMed] [Google Scholar]
- 78.Kozlov AG, Lohman TM. E. coli SSB tetramer binds the first and second molecules of (dT)(35) with heat capacities of opposite sign. Biophys Chem. 2011;159:48–57. doi: 10.1016/j.bpc.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kozlov AG, Lohman TM. Effects of monovalent anions on a temperature-dependent heat capacity change for Escherichia coli SSB tetramer binding to single-stranded DNA. Biochemistry. 2006;45:5190–5205. doi: 10.1021/bi052543x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kozlov AG, Lohman TM. Large contributions of coupled protonation equilibria to the observed enthalpy and heat capacity changes for ssDNA binding to Escherichia coli SSB protein. Proteins Suppl. 2000;4:8–22. doi: 10.1002/1097-0134(2000)41:4+<8::aid-prot20>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 81.Holbrook JA, Tsodikov OV, Saecker RM, Record MT., Jr Specific and non-specific interactions of integration host factor with DNA: Thermodynamic evidence for disruption of multiple IHF surface salt-bridges coupled to DNA binding. J Mol Biol. 2001;310:379–401. doi: 10.1006/jmbi.2001.4768. [DOI] [PubMed] [Google Scholar]
- 82.Bergqvist S, Williams MA, O'Brien R, Ladbury JE. Heat capacity effects of water molecules and ions at a protein-DNA interface. J Mol Biol. 2004;336:829–842. doi: 10.1016/j.jmb.2003.12.061. [DOI] [PubMed] [Google Scholar]
- 83.Yang Y, LiCata VJ. Interactions of replication versus repair DNA substrates with the Pol I DNA polymerases from Escherichia coli and Thermus aquaticus. Biophys Chem. 2011;159:188–193. doi: 10.1016/j.bpc.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 84.Datta K, Johnson NP, LiCata VJ, von Hippel PH. Local conformations and competitive binding affinities of single- and double-stranded primer-template DNA at the polymerization and editing active sites of DNA polymerases. J Biol Chem. 2009;284:17180–17193. doi: 10.1074/jbc.M109.007641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Filfil R, Chalikian TV. The thermodynamics of protein-protein recognition as characterized by a combination of volumetric and calorimetric techniques: The binding of turkey ovomucoid third domain to alpha-chymotrypsin. J Mol Biol. 2003;326:1271–1288. doi: 10.1016/s0022-2836(03)00022-6. [DOI] [PubMed] [Google Scholar]
- 86.Baker BM, Murphy KP. Dissecting the energetics of a protein-protein interaction: The binding of ovomucoid third domain to elastase. J Mol Biol. 1997;268:557–569. doi: 10.1006/jmbi.1997.0977. [DOI] [PubMed] [Google Scholar]
- 87.Armstrong KM, Insaidoo FK, Baker BM. Thermodynamics of T-cell receptor-peptide/MHC interactions: Progress and opportunities. J Mol Recognit. 2008;21:275–287. doi: 10.1002/jmr.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Frisch C, Schreiber G, Johnson CM, Fersht AR. Thermodynamics of the interaction of barnase and barstar: Changes in free energy versus changes in enthalpy on mutation. J Mol Biol. 1997;267:696–706. doi: 10.1006/jmbi.1997.0892. [DOI] [PubMed] [Google Scholar]
- 89.Sun D, Lopez-Guajardo CC, Quada J, Hurley LH, Von Hoff DD. Regulation of catalytic activity and processivity of human telomerase. Biochemistry. 1999;38:4037–4044. doi: 10.1021/bi982249n. [DOI] [PubMed] [Google Scholar]
- 90.Trakselis MA, Benkovic SJ. Intricacies in ATP-dependent clamp loading: Variations across replication systems. Structure. 2001;9:999–1004. doi: 10.1016/s0969-2126(01)00676-1. [DOI] [PubMed] [Google Scholar]
- 91.Kamtekar S, Berman AJ, Wang J, Lazaro JM, de VM, Blanco L, Salas M, Steitz TA. The phi29 DNA polymerase: Protein-primer structure suggests a model for the initiation to elongation transition. EMBO J. 2006;25:1335–1343. doi: 10.1038/sj.emboj.7601027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Blanco L, Bernad A, Lazaro JM, Martin G, Garmendia C, Salas M. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J Biol Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- 93.Beckman JW, Wang Q, Guengerich FP. Kinetic analysis of correct nucleotide insertion by a Y-family DNA polymerase reveals conformational changes both prior to and following phosphodiester bond formation as detected by tryptophan fluorescence. J Biol Chem. 2008;283:36711–36723. doi: 10.1074/jbc.M806785200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dionne I, Nookala RK, Jackson SP, Doherty AJ, Bell SD. A heterotrimeric PCNA in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol Cell. 2003;11:275–282. doi: 10.1016/s1097-2765(02)00824-9. [DOI] [PubMed] [Google Scholar]
- 95.Dionne I, Brown NJ, Woodgate R, Bell SD. On the mechanism of loading the PCNA sliding clamp by RFC. Mol Microbiol. 2008;68:216–222. doi: 10.1111/j.1365-2958.2008.06150.x. [DOI] [PubMed] [Google Scholar]
- 96.Gotz D, Paytubi S, Munro S, Lundgren M, Bernander R, White MF. Responses of hyperthermophilic crenarchaea to UV irradiation. Genome Biol. 2007;8:R220. doi: 10.1186/gb-2007-8-10-r220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Frols S, Gordon PM, Panlilio MA, Duggin IG, Bell SD, Sensen CW, Schleper C. Response of the hyperthermophilic archaeon Sulfolobus solfataricus to UV damage. J Bacteriol. 2007;189:8708–8718. doi: 10.1128/JB.01016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.