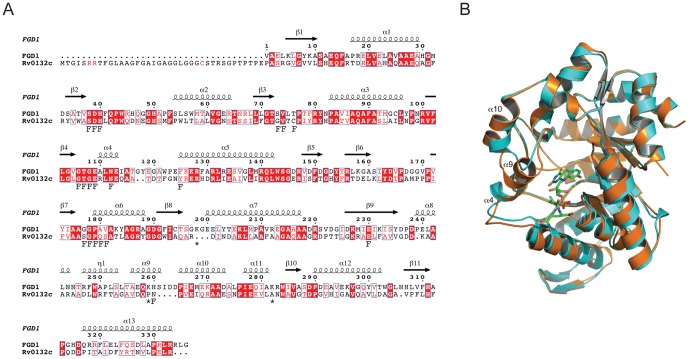
Figure 4. Structural comparison of Rv0132c with FGD1.
(A) Amino acid sequence alignment. The secondary structure elements for FGD1 [5] are shown above the sequence. FGD1 residues that hydrogen bond with F420 or the phosphate group of glucose-6-phosphate are indicated below the sequence by F and asterisk, respectively. The twin arginines in the Tat motif and the critical cysteine residue in the lipobox motif are shown in red in the Rv0132c signal sequence. (B) Superposition of the FGD1 (orange) crystal structure on the modeled Rv0132c (cyan). The F420 cofactor (green) bound to FGD1 is shown in stick representation. Replacement of helix α9 with a smaller loop extends the active site cavity in Rv0132c. For details of FGD1 structure see [5].