Abstract
Centrioles are duplicated during S-phase to generate the two centrosomes that serve as mitotic spindle poles during mitosis. The centrosomal pool of the Mps1 kinase is important for centriole assembly, but how Mps1 is delivered to centrosomes is unknown. Here we have identified a centrosome localization domain within Mps1 and identified the mitochondrial porin VDAC3 as a protein that binds to this region of Mps1. Moreover, we show that VDAC3 is present at the mother centriole and modulates centriole assembly by recruiting Mps1 to centrosomes.
Keywords: Mps1, VDAC, centriole assembly, centrosome, kinase
Introduction
Centrosomes consist of a pair of centrioles that must be precisely duplicated during S-phase to ensure bipolar spindle assembly and maintenance of genomic integrity in the progeny.1 Of the two centrioles, the “mother” is at least one cell cycle older than the other, its daughter, and is distinguishable by the presence of appendages.2 Around the G1/S transition, each centriole assembles a procentriole at its proximal end that gradually elongates to form a mature centriole by mitosis.3 At the base of each procentriole, Sas6 forms the central hub of a “cartwheel” that establishes the 9-fold symmetry of the centriole4,5 and behaves as the scaffold onto which other centriolar proteins are assembled during centriole assembly.3,6 During quiescence, the mother centriole is converted to a basal body to organize a primary cilium that is resorbed during cell cycle re-entry.7
The Mps1 protein kinase is essential for the spindle assembly checkpoint8 and modulates centriole assembly in vertebrates.9–13 While recent studies suggest Mps1 may be dispensable for centriole assembly,6 Mps1 is nonetheless an important regulator of centriole assembly. Mps1 phosphorylates Cetn2 and is required for its recruitment to procentrioles.11 Moreover, increasing the level of Mps1 at centrosomes causes centriole over-duplication in every human cell type thus far tested.10,12–15 Because extra centrosomes are implicated in tumorigenesis,1,14–16 this makes it important to understand the mechanisms that regulate centrosomal Mps1, even if centrioles can be built in its absence. However, while centrosomal Mps1 levels are fine-tuned by proteasome-mediated degradation,10,12 the mechanisms that target Mps1 to centrosomes are not known.
In this study, we have characterized a centrosome localization domain (CLD) in Mps1 and using yeast two-hybrid identified voltage dependent anion channel 3 (VDAC3) as a CLD-binding protein. VDAC3 belongs to the mitochondrial porins, but while it shares 60–70% sequence identity with VDAC1 and VDAC2, which regulate mitochondrial energetics,17,18 the mitochondrial function of VDAC3 is not clear. Moreover, VDAC3 has been found at the non-membranous sperm outer dense fiber, where the maternal centriole protein ODF2 was first identified.19,20 We show here that in addition to interacting with Mps1 in vitro and in vivo, VDAC3 is also a centrosomal protein of the maternal centriole that regulates centriole assembly by recruiting Mps1 to the centrosome.
Results
Mps1 physically interacts with VDAC3
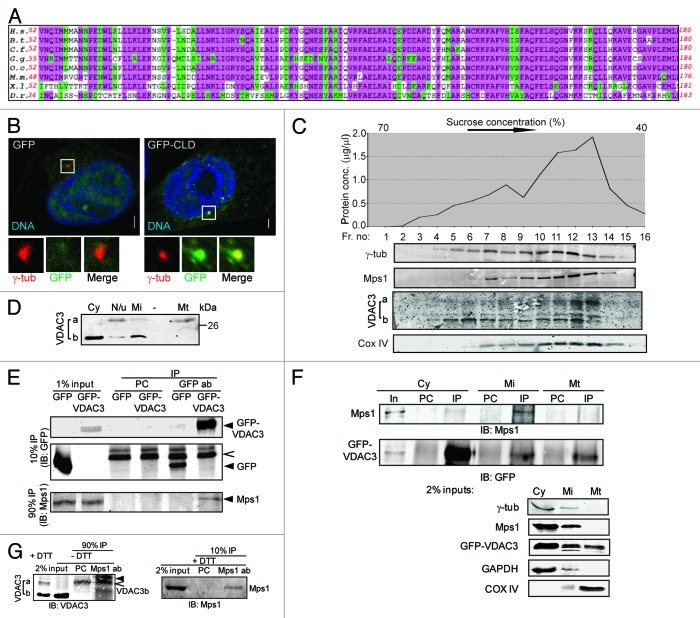
Consistent with its key roles in spindle assembly checkpoint and centrosome assembly, Mps1 is found at kinetochores in mitosis and centrosomes at all points in the cell cycle. 9,21 We noticed that Mps1 residues 52–180 are highly conserved among vertebrates (Fig. 1A), and reasoning that Mps1 must contain a centrosomal targeting motif, wondered if this region might serve to target Mps1 to the centrosome. We found that Mps153-175 directs GFP to centrosomes in roughly 30% of HeLa (Fig. 1B), RPE1 (Fig. S2D) and NIH3T3 cells (data not shown), while GFP alone was only rarely found at centrosomes in cells with comparable fluorescence (Fig. 1B). Because this region is sufficient to direct GFP to centrosomes, we refer to it as the Mps1 centrosome localization domain (CLD). The Mps1 N terminus also directs kinetochore localization through a tetratricopeptide repeat motif,21,22 and thus mediates diverse sub-cellular localization through binding with different sets of scaffold proteins.
Figure 1. VDAC3 physically interacts with Mps1. (A) Conservation in the Mps1 N terminus; highlighted residues are identical (pink) or conserved in structure or function (green) in at least 75% of the species. H.s., Homo sapiens (human); B.s., Bos taurus (bovine); C.f., Canis familiaris (dog); G.g., Gallus gallus (chicken); O.c., Oryctolagus cuniculus (European rabbit); M.m., Mus musculus (mouse); X.l., Xenopus laevis (frog); D.r., Danio rerio (zebrafish). (B) S-phase arrested HeLa cells expressing GFP or GFP-Mps153-175 (GFP-CLD) were stained for γ-tubulin (γ-tub; red) and DNA (blue). Bar = 5 μm. In this and other figures, panels show 4-fold digitally magnified images of a region of interest, in this case surrounding the centrosomes. (C) Sucrose gradient fractionation of nucleui-depleted extract from HeLa cells, where the graph represents the total protein concentrations of the fractions and the immunoblots show the distribution of indicated protein in these fractions. The absence of nuclear protein contamination was verified by immunoblotting with anti-LaminB antibody (data not shown). (D) Differential distribution of VDAC3a and VDAC3b as shown by immunoblotting of subcellular fractions [cytosolic (Cy), mitochondrial (Mt), microsomal (Mi) and pellet (N/u)] obtained using Qproteome mitochondria isolation protocol. (E) After pre-clearing with beads alone (PC), anti-GFP immunoprecipitates from S-phase arrested HEK293 cells expressing GFP or GFP-VDAC3 (arrowheads) were immunoblotted with antibodies against Mps1 and GFP. Caret indicates Protein G. (F) After pre-clearing (PC), GFP-VDAC3 immunoprecipitates (IP) from the subcellular fractions from S-phase arrested HEK293 cells expressing GFP-VDAC3 were immunoblotted as in (D). Either 1% (cytosol) or 2% (other fractions) of the input (In) were analyzed with the indicated antibodies. Although a truncated form of GFP-VDAC3 was detected in lysates of cells expressing GFP-VDAC3 (see Fig. S1F), only the band of expected size (full-length GFP-VDAC3) was shown for convenience, in (E and F). (G) After pre-clearing (PC), anti-Mps1 immunoprecipitates from S-phase arrested RPE1 cells were separated in reducing (+DTT) or non-reducing (-DTT) condition and immunoblotted with indicated antibodies. Trace amount of Protein G and IgG light chain are indicated by caret and arrowhead, respectively.
To identify proteins that bind to the CLD, we screened a human cDNA library against the CLD by yeast two-hybrid (see Materials and Methods). One positive clone contained the 3′-region of VDAC3. This appeared to reflect a true physical interaction, because recombinant GST-CLD binds to GFP-VDAC3 expressed in HEK293 cells but not to GFP alone (Fig. S1A). Conversely, recombinant MBP-VDAC3 binds to endogenous Mps1 from cell lysates (Fig. S1B). While VDAC1 and VDAC2 control bioenergetics and mitochondrial-mediated apoptosis,23 the mitochondrial function of VDAC3 is unclear.24 Additionally, both VDAC2 and VDAC3 were identified in the sperm outer dense fiber,19 a non-membranous structure associated with the sperm flagella where the maternal centriole protein ODF2 was first found,20 and the fly porin was identified in a proteomic study of Drosophila centrosomes,25 encouraging us to pursue the interaction between Mps1 and VDAC3.
In this study, we used two commercially available antibodies directed against overlapping peptides derived from the VDAC3 N terminus. These antibodies behaved identically in all assays described herein, and for convenience we refer to them as a single antibody that recognizes the N terminus of VDAC3 (Fig. S1C) and does not significantly cross react with VDAC1 or VDAC2 (Fig. S1D). Interestingly, in addition to the expected band at 30 kDa (hereafter VDAC3a), we observed a stronger band of 24 kDa (hereafter VDAC3b) from the lysates of various human cells, including HeLa (Figs. 1C and D and 5F), RPE1 (Figs. 1G and 3D), HEK 293 and U2OS (data not shown). An alternatively spliced VDAC3 message containing a single extra ATG (118ATG) that creates a putative alternative translational start site for a protein with the approximate size of VDAC3b has been described.26 However, we were able to detect only a very small amount of this message in RPE1 cells, where the most abundant VDAC3 message lacks 118ATG (Fig. S1E). Moreover, an antibody against GFP detects two species in lysates from cells transfected with a GFP-VDAC3 cDNA, one at the predicted size and a second at ~30 KDa that is consistent with the size of GFP plus a portion of the VDAC3 N terminus (Fig. S1F and G). Because the smaller form includes GFP, it cannot be derived from a translational start site within the VDAC3 coding sequence. The proteasome inhibitor MG115 does not alter the abundance of the smaller form (Fig. S1F), suggesting that it is not a degradation product and both GFP-positive species were downregulated (Fig. S1G) by a VDAC3-specific siRNA (see below). Although elucidation of the precise mechanism for the presence of two VDAC3 isoforms is beyond the scope of this study, these observations suggest that VDAC3a and VDAC3b are derived from the same message, and that VDAC3b arises through a post-translational mechanism.
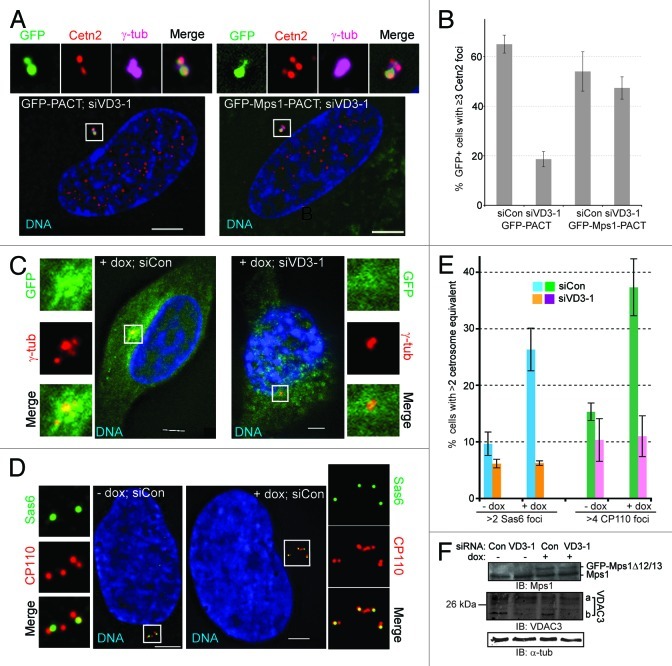
Figure 5. VDAC3 recruits Mps1 to centrosomes to regulate centriole assembly. (A and B) siCon or siVDAC3–1 (siVD3–1) RPE1 cells expressing GFP-PACT (green) or GFP-Mps1-PACT (green) were arrested in S-phase by a 24 h HU treatment and were examined for centriole numbers. (A) Shown are representative siVDAC3–1 cells stained for Cetn2 (red), γ-tub (magenta) and DNA (blue); GFP is green, bar = 5 μm. (B) Percentage of GFP-positive cells with ≥ 3 Cetn2 foci, values represent the mean ± SD for three independent experiments, 50–60 cells counted per replicate. (C–F) HeLa GFP-Mps1∆12/13 cells transfected with control (siCon) and VDAC3–1 (siVD3–1) siRNA were arrested in S-phase by a 72 h HU treatment, with or without dox, to induce GFP-Mps1∆12/13 [green in (C)], and stained for DNA (blue) and (C) γ-tub (red) or (D) CP110 (red) and Sas6 (green). Bar = 5 μm. (E) The percentage of cells with more than two centrosome equivalents (> 2 Sas6 foci or > 4 CP110 foci), values represent the mean ± SD for three independent experiments, at least 100 cells counted per replicate. (F) Immunoblots showing the expression of GFP-Mps1∆12/13 (arrowhead), endogenous Mps1 (arrow) and depletion of VDAC3 (VDAC3a roughly 10%, VDAC3b roughly 80%), α-tub as loading control.
Figure 3. VDAC3 recruits Mps1 to centrosomes. (A and B) Representative RPE1 cells stained for Mps1 (green) and (A) Ninein (red) or (B) VDAC3 (red). DNA is blue, bar = 5 μm. (C and D) Asynchronously growing RPE1 cells transfected with control (Con) or VDAC3 siRNA-1 (VD3–1) analyzed by (C) qRT-PCR or (D) immunoblot. (C) Relative expression of VDAC3 mRNA, values represent mean ± SD for three replicates. (D) Immunoblot showing VDAC3a decreased by roughly 35%, and VDAC3b by 75%. GAPDH was used as loading control. (E and F) Representative images of RPE1 cells prepared as in (C and D) stained for VDAC3 (red) and (E) Ac-tub (green) or (F) Cep170 (green). DNA is blue, bar = 5 μm. Panels show magnified centrosomes/basal bodies. In (F), cartoon shows localization of VDAC3 (red) with respect to Cep170 (green) in control and VDAC3-depleted RPE1 cells. (G) Micrographs show representative images of RPE1 cells prepared as in (C and D), treated with DMSO or MG115, labeled with BrdU for 4 h and stained for Mps1 (green), γ-tub (red) and BrdU (blue). Panels show magnified centrosomes. Bar = 5 μm. Immunoblots show whole-cell level of Mps1, α-tubulin (α-tub) as loading control. Centrosomal Mps1 level was determined as described in Materials and Methods. Values represent mean ± SD of 25 representative cells.
After sucrose gradient centrifugation of cellular proteins, VDAC3a co-fractionated with the mitochondrial marker COX IV, whereas VDAC3b was found in high-density fractions that contained γ-tubulin and Mps1 (Fig. 1C). Consistent with this observation, VDAC3a is predominantly present in the mitochondrial fraction after subcellular fractionation, while VDAC3b is absent from the mitochondrial fraction and is instead found in the cytosolic and microsomal fractions (Fig. 1D). Although the VDAC3 antibody could not successfully immunoprecipitate endogenous VDAC3, a small fraction of Mps1 co-immunoprecipitates with GFP-VDAC3 from the microsomal fraction, which was depleted of mitochondria but contained both γ-tubulin and Mps1 (Fig. 1E and F). Notably, this co-immunoprecipitation was not seen in the mitochondrial fraction (Fig. 1F), demonstrating that it is a non-mitochondrial pool of GFP-VDAC3 that interacts with Mps1. Mps1 also interacts with endogenous VDAC3, as a fraction of VDAC3 co-immunoprecipitates with Mps1 (Fig. 1G). We assume that the interacting species is VDAC3b based on its size, but note that it migrated slightly more slowly than VDAC3b from the input. This could be due to the sample handling required to minimize interference from IgG light chain (caret in Fig. 1G), but it is also possible that Mps1 preferentially interacts with a modified form of VDAC3. Because VDAC3 is close in size to both IgG light chain and Protein G (Fig. 1F), and because the VDAC3 antibody did not work for immunoprecipitation, we did not pursue further co-immunoprecipitation studies. Regardless, these observations support the existence of both soluble and mitochondrial pools of VDAC3, and show that VDAC3 co-fractionates with centrosomes and physically interacts with Mps1.
VDAC3 localizes to centrosomes
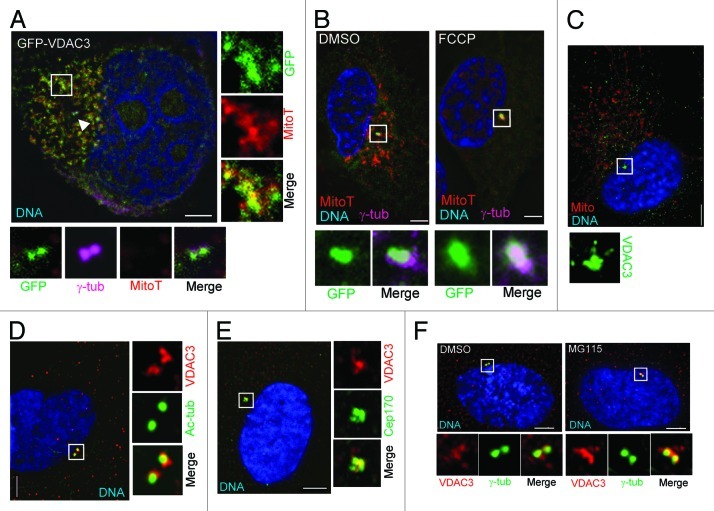
If the VDAC3-Mps1 interaction described above is relevant to the centrosomal pool of Mps1, VDAC3 should be present at centrosomes in addition to its well-known mitochondrial localization. Accordingly, we next sought to characterize the subcellular localization of VDAC3. Consistent with our subcellular fractionation data, we found that GFP-VDAC3 was both diffusely cytosolic and found in punctate structures that co-localized with mitochondria in human cells including HeLa and RPE1 cells (Fig. 2A and B). We also found that GFP-VDAC3 was localized to centrosomes, as judged by its co-localization with γ-tubulin in 30–35% and 45–50% of asynchronously growing and S-phase arrested HeLa cells, respectively, and roughly 60% of asynchronous RPE1 cells (Fig. 2A and B). This centrosomal localization is not due to GFP at its N terminus, as DsRed-VDAC3 (Fig. S2A), and C-terminally tagged VDAC3-GFP (data not shown) showed similar localization. A 1 h treatment with FCCP, an uncoupler of oxidative phosphorylation that depolarizes the mitochondrial membrane and abolishes the microtubular network (see below and ref. 27), had no effect on centrosomal GFP-VDAC3, despite diminishing both cytosolic and mitochondrially localized GFP-VDAC3 as well as Mitotracker labeling (Fig. 2B).
Figure 2. VDAC3 is a centrosomal protein of the mother centriole. (A) A representative HeLa cell expressing GFP-VDAC3 (green) stained with MitotrackerRed (MitoT, red), then fixed and stained for γ-tub (magenta). Arrowhead indicates centrosomes, box indicates a region containing mitochondria. (B) RPE1 cells expressing GFP-VDAC3 (green) were treated with either DMSO (solvent control) or 200 μM FCCP for 1 h and stained with MitoT (red). Shown are representative images stained for γ-tub (magenta). (C–E) Asynchronously growing RPE1 cells were stained for (C) VDAC3 (green) and mitochondria (Mito, red), (D) VDAC3 (red) and Ac-tubulin (green), and (E) VDAC3 (red) and Cep170 (green). Shown are representative images where the predominant centrosomal VDAC3 signal was at both centrosomes (C and D) or at the mother centriole (E). (F) Representative images of asynchronously growing RPE1 cells treated with either DMSO or MG115 for 4 h, stained for γ-tub (green), VDAC3 (red) and imaged under identical condition. DNA is blue and bar is 5 μm in (A–F).
The VDAC3 antibodies described above only weakly stained mitochondria (Fig. 2C) but stained centrosomes comparatively strongly in RPE1 (Figs. 2C–F and 3B) and HeLa cells (Fig. S2B). This centrosomal signal was blocked by pre-incubation of the antibodies with recombinant VDAC3 (Fig. S2B) and, thus, was not background staining. Though less apparent in HeLa cells, centrosomal VDAC3 was always more strongly associated with one of the two centrosomes (Fig. 2D–E; Fig. 2F, DMSO panel; Fig. S2C), and in roughly 70% of RPE1 cells, the bulk of VDAC3 lay within the region defined by Cep170 (Fig. 2E; Fig. S3A), a centriolar appendage marker.28 These observations suggest the presence of a centrosomal pool of VDAC3 that is preferentially associated with the maternal centriole. We attempted to verify this ultrastructurally, but the VDAC3 antibody failed to generate a signal in immuno-EM, either at mitochondria or centrioles.
The accumulation of VDAC3 at centrosomes varies with the cell cycle. GFP-VDAC3 was found to be centrosomal in roughly 20% of cells with a single centrosome that were presumably in G1; 60% of cells with two closely spaced centrosomes that were presumably in S-phase; and 20% of cells with two separated centrosomes that were presumably in G2 or mitosis. The intensity of endogenous VDAC3 also varies through the cell cycle, with the maximum level of centrosomal VDAC3 present during S-phase (as identified by two closely spaced centrosomes) and a substantial reduction in the level during mitosis (Fig. S2C). Interestingly, DsRed-VDAC3 was intensely centrosomal in ~99% of cells after a 4 h treatment with MG115, which had no effect on DsRed alone or the whole-cell level of DsRed-VDAC3 (Fig. S2A). The centrosomal signal for endogenous VDAC3 was similarly increased in intensity upon MG115 treatment and was also expanded in extent, being distributed over both the centrosomes, as compared with DMSO-treated cells (Fig. 2F). In parallel with these changes in VDAC3, MG115 increased the percentage of cells in which GFP-CLD accumulated at centrosomes to 63 ± 5%, as compared with 27 ± 3% in DMSO-treated cells, supporting the suggestion that centrosomal VDAC3 levels affect the recruitment of Mps1 to centrosomes. These observations suggest that like that of Mps1,12 the centrosomal pool of VDAC3 may be regulated by degradation, perhaps in a cell cycle-dependent manner.
VDAC3 recruits Mps1 to centrosomes
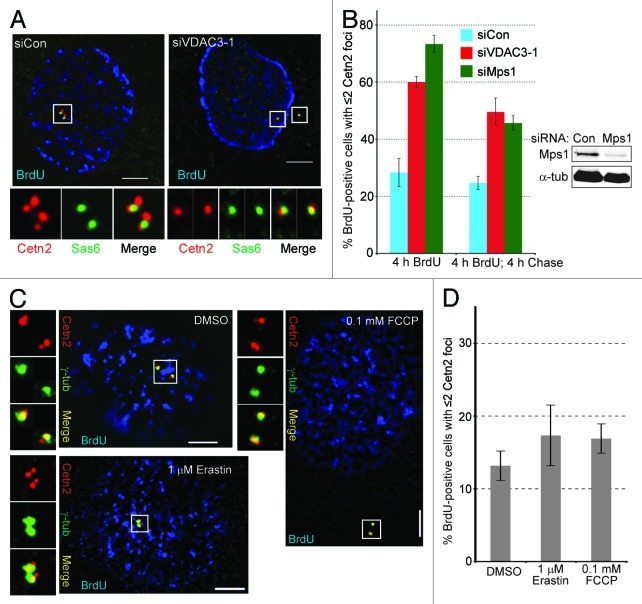
In order to test the hypothesis that VDAC3 recruits Mps1 to centrosomes, we examined centrosomal Mps1 after the siRNA-mediated depletion of VDAC3. While the Mps1 antibody used here stains both centrosomes in HeLa cells,10 in RPE1 cells it almost exclusively stained the Ninein-positive centrosome (Fig. 3A) where it overlapped significantly with VDAC3 (Fig. 3B), suggesting that in RPE1 cells, Mps1 predominantly localizes to the maternal centriole. Compared with a control siRNA (siCon), a VDAC3-specific siRNA (siVDAC3–1) reduced the VDAC3 transcript by 70–75% (Fig. 3C). It also reduced VDAC3 protein levels by 65–70% (VDAC3b) and 30–35% (VDAC3a) (Fig. 3D) and exogenously expressed GFP-VDAC3 by 70% (Fig. S1G) compared with control cells. Interestingly, centrosomal VDAC3 was not completely lost in siVDAC3 cells, although its intensity was diminished, and its distribution was significantly altered; VDAC3 remained at daughter centrioles, but was lost from the maternal centriole/basal body (Fig. 3E) and was found on the periphery of the Cep170 signal (Fig. 3F; Fig. S3A). This change in centriolar VDAC3 staining is consistent with the level of depletion we achieved, and there are several examples of centriolar proteins that are not completely depleted from centrioles by siRNA, including POC5,29 Cep15230 and Cetn2.31 This residual VDAC3 staining in siVDAC3 cells is not background staining based on the complete loss of centrosomal staining when the antibody is pre-incubated with recombinant VDAC3 (Fig. S2B), and might reflect VDAC3a, which was less well depleted than VDAC3b. While it is formally possible that the VDAC3 antibody cross reacts with another centriolar protein, we consider this possibility unlikely, because the putative cross-reactive protein would have to show cell cycle- and proteasome-dependence similar to that of GFP-VDAC3 and depend on VDAC3 for its centrosomal accumulation.
While γ-tubulin staining at the centrosome was unaltered in siVDAC3 cells, centrosomal Mps1 was greatly reduced (Fig. 3G). To quantify this reduction, we modified our quantitative microscopy approach10 to normalize the centrosomal Mps1 signal to that of γ-tubulin (see Materials and Methods). To rule out any effect of cell cycle position, we restricted our analysis to cells that were in S-phase as judged by incorporation of BrdU during a 4 h labeling and had two centrosomes as judged by γ-tubulin staining. The average normalized Mps1 signal at centrosomes was roughly 2.5-fold lower in BrdU-positive siVDAC3 cells compared with BrdU-positive siCon cells (Fig. 3G). This decrease is not due to the previously described degradation of centrosomal Mps1,12 because MG115 had no significant effect on centrosomal Mps1 in siVDAC3 cells, despite causing a 2-fold increase in siCon cells (Fig. 3G). Unlike HeLa cells,12 MG115 caused a 25–30% increase in whole-cell Mps1 in RPE1 cells (Fig. 3G), suggesting that the recently described cytoplasmic degradation of Mps1 in telophase and G115 is more robust in RPE1 cells. This increase was not seen in siVDAC3 cells, which likely reflects a cell cycle effect due to the large percentage of siVDAC3 cells that fail to incorporate BrdU (see below). Expression of a siRNA-resistant version of VDAC3 (GFP-sirVDAC3) in siVDAC3 cells increased the centrosomal level of Mps1 compared with adjacent GFP-negative siVDAC3 cells (Fig. S3B), demonstrating that the reduction of centrosomal Mps1 was specific to the depletion of VDAC3. A similar reduction in centrosomal Mps1 level was also seen in RPE1 cells treated with a second siRNA against VDAC3 (siVDAC3–2, Fig. S3C), further suggesting that the phenotype is an effect of VDAC3 knockdown rather than a non-specific or off target effect of VDAC3 siRNA. Together, these results suggest that VDAC3, alone or in a complex, regulates the recruitment of Mps1 to centrosomes.
The failure to recruit Mps1 to centrosomes inhibits centriole assembly in siVDAC3 cells
If VDAC3 recruits Mps1 to centrosomes, VDAC3 depletion should attenuate centriole assembly in a manner similar to that of Mps1. To test this prediction, we examined centrioles in cells that had entered S-phase as judged by BrdU incorporation. Neither siVDAC3 nor an Mps1-siRNA (siMps1) prevented procentriole assembly, because all BrdU-positive siVDAC3 and siMps1 cells had two Sas6 foci (Fig. 4A). However, both siVDAC3 and siMps1 prevented efficient incorporation of centriolar proteins into procentrioles, as judged by an elevated percentage of BrdU-positive cells with two foci of Cetn2 (Fig. 4A and B) or CP110 (Fig. S4A and B) after a 4 h BrdU pulse, with a larger effect on Cetn2. The percentage of cells with two Cetn2 or CP110 foci dropped after a 4 h chase (Fig. 4B; Fig. S4B), suggesting that in some cells, the incorporation of Cetn2 and CP110 was merely delayed, as we recently found in Cetn2-depleted HeLa cells.11 However, the percentage of BrdU-positive cells with two Cetn2 foci remained at 45–50% even after the 4 h chase (Fig. 4B), suggesting that many siVDAC3 and siMps1 cells were unable to assemble complete centrioles. Interestingly, only ~10% of siVDAC3 or siMps1 cells incorporated BrdU during a 4 h pulse, compared with 40–45% of siCon cells. While the reason for this is unclear, it seems to be an effect on cell cycle entry rather than progression, because all BrdU-positive siVDAC3 and siMps1 cells had two Sas6 foci, demonstrating that there is no delay in the initiation of centriole assembly upon S-phase entry in siVDAC3 cells. Therefore, because we limited our analysis to S-phase cells, all of which had initiated centriole assembly, it is unlikely that the defects we observed in incorporation of Cetn2 and CP110 into centrioles are secondary to any cell cycle defect. Consistent with our observations in asynchronous cells, siVDAC3 caused a 3-fold reduction in the percentage of cells with more than two Cetn2 foci in hydroxyurea (HU)-arrested cells expressing GFP (Fig. S5A and B). However, expression of GFP-sirVDAC3 restored centriolar incorporation of Cetn2 in HU-arrested siVDAC3 cells to levels comparable to that seen in siCon cells (Fig. S5A and B). Because cells that had not entered the cell cycle would not have replicated their centrioles, this demonstrates that both the centriole assembly defect and the cell cycle defect are specific to the depletion of VDAC3.
Figure 4. Depletion of VDAC3 or Mps1 inhibits centriole assembly. (A and B) Asynchronously growing RPE1 cells transfected with control (siCon), VDAC3- (siVDAC3–1) or Mps1-specific (siMps1) siRNAs were labeled with a 4 h pulse of BrdU. Centriole number was determined after this pulse (4 h BrdU), or after an additional 4 h chase (4 h BrdU, 4 h chase) using antibodies against Cetn2 and Sas6. (A) Representative images of siCon or siVDAC3–1 cells stained for Cetn2 (red), Sas6 (green) and BrdU (blue). Bar = 5 μm. (B) Percentage of BrdU-positive cells with ≤ 2 Cetn2 foci. Values represent mean ± SD for three independent experiments, 75–100 cells counted per replicate. Immunoblots show depletion of Mps1 (by roughly 85%), α-tub as loading control. (C and D) Growing RPE1 cells were treated with DMSO, 1 μM Erastin for 16 h or 100 μM FCCP for 4 h, labeled with a 4 h pulse of BrdU and stained for Cetn2. (C) Shown are BrdU (blue), Cetn2 (red) and γ-tub (green) for representative cells. Bar = 5 μm. (D) Percentage of BrdU-positive cells with ≤ 2 Cetn2 foci, values represent mean ± SD for three independent experiments, 75–100 cells counted per replicate.
If the effect of VDAC3 depletion on centriole assembly were an indirect effect of inhibiting a mitochondrial function of VDAC3, inhibiting mitochondrial VDAC3 or perturbing mitochondrial function in general should have the same effect. However, neither Erastin, an inhibitor of the mitochondrial function of VDAC3 and VDAC2,32,33 nor FCCP altered the number of Cetn2 foci in BrdU-positive cells (Fig. 4C and D). Because the compounds had the expected effect on cell viability and microtubules (Fig. S4C and D), and because neither GFP-VDAC3 nor endogenous VDAC3 could be observed at mitochondria in FCCP-treated cells (see above), this suggests that the centriole assembly defects in siVDAC3 cells do not reflect inhibition of a mitochondrial function of VDAC3 or a general effect of mitochondrial dysfunction. In contrast, if the centriole assembly defect in siVDAC3 cells is due to the failure to recruit Mps1 to centrosomes, it should be suppressed by tethering Mps1 to centrosomes independently of VDAC3, e.g., via the PACT domain.12,34 As with GFP alone, siVDAC3 caused a 3-fold reduction in the percentage of HU-arrested cells expressing GFP-PACT that had more than two Cetn2 foci (Fig. 5A and B; Fig. S5C). However, siVDAC3 did not cause any decrease in the percentage of cells with more than two Cetn2 foci in HU-arrested cells expressing GFP-Mps1-PACT (Fig. 5A and B; Fig. S5C), an exclusively centrosomal version of Mps1 that is functional in centriole assembly.12 Interestingly, this observation demonstrates that GFP-Mps1-PACT also suppresses the cell cycle defect caused by siVDAC3, because cells that had not entered the cell cycle would not have replicated their centrioles, suggesting that the role of VDAC3 in both cell cycle and centriole assembly may be related to centrosomal Mps1. The observations that the centriole assembly defect in siVDAC3 cells is specific to the depletion of VDAC3, is not caused by the VDAC3-inhibitor Erastin, and can be suppressed by GFP-Mps1-PACT support the hypothesis that a centriolar pool of VDAC3 regulates centriole assembly by recruiting Mps1 to centrosomes.
Mps1-dependent centriole reduplication requires VDAC3
If VDAC3 recruits Mps1 to centrosomes, VDAC3 depletion should attenuate Mps1-dependent centriole over production. To test this, we utilized HeLa-GFP-Mps1∆12/13 cells.14 In the presence of doxycycline (dox), these cells express low levels of GFP-Mps1∆12/13, which drives centriole re-duplication because it cannot be properly degraded at centrosomes.12,14 In the presence of dox, a significant percentage of siCon cells had more than two foci of γ-tubulin or Sas6 or more than four foci of CP110 (Fig. 5C–E). However, VDAC3 depletion markedly restricted this centriole reduplication (Fig. 5D–F). The siVDAC3 and siCon cells had entered the cell cycle and were arrested in S-phase by HU, because they all had at least two Sas6 foci. Moreover, unlike RPE1 cells, siVDAC3 does not cause any apparent defect in cell cycle entry in HeLa cells. Therefore, the failure to undergo centriole re-duplication is not the consequence of a cell cycle delay. Because GFP-Mps1∆12/13 cannot be degraded at centrosomes,12 this further supports the suggestion that Mps1 cannot bind to centrosomes in the absence of VDAC3. Indeed, it was difficult to detect GFP-Mps1∆12/13 in the vicinity of centrosomes in siVDAC3 cells (Fig. 5C), although we did not quantify this effect. This observation also demonstrates that in the absence of the PACT domain, Mps1 cannot suppress the effects of VDAC3 depletion.
Discussion
We have found that VDAC3 interacts with the centrosomal protein Mps1 and localizes to centrosomes, where it preferentially associates with the mother centriole. Consistent with the hypothesis that VDAC3 recruits Mps1 to centrosomes, Mps1 is lost from centrosomes in VDAC3-depleted cells. This loss cannot be reversed by proteasome inhibition, suggesting it reflects a failure to recruit Mps1 to centrosomes rather than a defect in stability at centrosomes. VDAC3-specific siRNAs cause a centriole assembly defect that is specific to the depletion of VDAC3, is similar to that in Mps1-depleted cells, is not due to inhibition of the mitochondrial function of VDAC3 and is bypassed by targeting Mps1 to centrosomes independently of VDAC3. Thus, our data strongly suggest that a centrosomal pool of VDAC3 modulates centriole assembly by recruiting Mps1 to the centrosome.
Perhaps the association of a mitochondrial porin with centrosomes should not be surprising. Even though no membranes are thought to exist at centrosomes, both membrane and mitochondrial proteins have been found at centrosomes and basal bodies.35–38 Notably, Mps1 binds to mortalin, which localizes to centrosomes, activates Mps1 kinase activity and is required for the ability of Mps1 to accelerate centrosome re-duplication despite also localizing to mitochondria.39 Moreover, VDAC2 and VDAC3 were found in the non-membranous sperm outer dense fiber19 and, thus, are not obligate membrane proteins, and VDAC2 was recently found at the base of primary cilia.35 VDAC1 auto inserts into artificial phospholipid bilayers to form a 19-stranded β barrel with a membrane-exposed face and an aqueous channel,40,41 and it is presumed this structure is representative of other VDACs.24 Such amphipathic β sheets can exist in both soluble and membrane-bound conformations,42 and indeed our data demonstrate the existence of a soluble pool of VDAC3b. This soluble pool of VDAC3b fractionates distinctly from mitochondria in sucrose gradients and, instead, co-fractionates with the high-density peak of γ-tubulin, which corresponds to the position of centrosomes.43 VDAC3b is also found in a subcellular fraction that is depleted of mitochondria but contains the centrosomal proteins γ-tubulin and Mps1. Thus, we hypothesize that centrosomal VDAC3 adopts a conformation where its hydrophobic surfaces are either buried or associated with hydrophobic surfaces of other centrosomal proteins.
Interestingly, we have also documented the presence of multiple isoforms of VDAC3. Our in vitro binding and co-immunoprecipitation data demonstrate that Mps1 can interact with full-length VDAC3 (e.g., MBP-VDAC3 binds to endogenous Mps1 and Mps1 co-immunoprecipitates with full-length GFP-VDAC3), but only VDAC3b co-immunoprecipitates with Mps1. Our data similarly show that neither the solubility nor the centrosome localization of VDAC3 is unique to VDAC3b, because full-length GFP-VDAC3 is present in the cytoplasmic fraction and VDAC3-GFP localizes to centrosomes. Therefore, we hypothesize that while Mps1 can interact with either form of VDAC3, it only interacts with VDAC3b in vivo, because that is the predominant form of endogenous VDAC3 present at centrosomes. Our study opens several interesting questions regarding the nature of VDAC3b, the VDAC3 isoform that co-fractionates with centrosomes. While it was ultimately concluded that the 118ATG VDAC3 message does not produce a truncated protein,44 that conclusion was based on analysis of mitochondria, and our observation that VDAC3b is not mitochondrial might suggest the need to reconsider that conclusion. However, we have made two observations suggesting that the 118ATG message does not account for VDAC3b; the level of the 118ATG message in RPE1 cells is too low to account for the more abundant VDAC3b, and a GFP-VDAC3 cDNA generates a truncated GFP-positive product that could not have arisen from translational initiation within VDAC3. Together, these observations suggest that VDAC3b arises through post-translational modification of a protein translated from the VDAC3 message, presumably VDAC3a. How VDAC3 gets to centrosomes, and what structure it adopts there, are also intriguing. However, these exciting questions are beyond the scope of this current study.
Our data demonstrate that depletion of VDAC3 leads to a defect in cell cycle entry. However, because we limited our analysis of asynchronous cells to those that had entered the cell cycle and initiated centriole assembly, the defects we observed in centriole assembly are not caused by this cell cycle entry defect. Interestingly, depletion of Mps1 caused a similar cell cycle defect, and tethering Mps1 to centrosomes via the PACT domain suppressed both the centriole assembly and cell cycle defects of siVDAC3. This suggests that the cell cycle defect in siVDAC3 cells may be related to centrosomal Mps1. Because we did not observe this cell cycle defect in HeLa cells, it is likely that it reflects the ability of RPE1 cells to enter G0. However, the reasons that cells with depleted centrosomal Mps1 might fail to enter the cell cycle are not yet clear. Depletion of several centrosomal proteins has been shown to lead to cell cycle arrest.45 While this was initially interpreted as a requirement for an intact centrosome in cell cycle progression, it has more recently been suggested that this arrest reflects a p38-dependent stress response.45
Although Mps1 may be dispensable for the canonical centriole duplication cycle, it plays an important role in recruiting Cetn2 during centriole maturation, and increasing the centrosomal Mps1 pool causes centriole reduplication. We have shown here that both functions of Mps1 require VDAC3. Taken together, our data demonstrate that VDAC3 is a novel centrosomal protein that recruits Mps1 to centrosomes to regulate centriole assembly. This raises many interesting questions, including how VDAC3 itself is targeted to centrosomes, the conformation of VDAC3 at centrosomes and whether other VDACs have similar centrosomal functions, which we look forward to addressing in future studies.
Materials and Methods
Plasmids
Previously described plasmids used for this study are- pHF7 (pECE-GFP) and pHF252 (pECE-DsRed).9,10,12 Plasmids created for this study are as follows: yeast two-hybrid bait plasmid pHF269 (Gal4BD-Mps153-175); bacterial expression constructs in the pDEST17 vector (Invitrogen), pHF270 (6xHis-VDAC1), pHF271 (6xHis-VDAC2), pHF272 (6xHis-VDAC3); in the pKM596 vector (Addgene), pHF273 (MBP-VDAC3); in the pGex6P-1 vector (GE Healthcare), pHF274 (GST-VDAC31-283), pHF275 (GST-VDAC31-136), pHF276 (GST-VDAC3181-283), pHF277 (GST-Mps150-175); mammalian expression constructs in the pECE vector, pHF278 (GFP- Mps153-175), pHF279 (GFP-VDAC3), pHF280 (VDAC3-GFP), pHF281 (GFP-VDAC3-PACT), pHF282 (DsRed-VDAC3), pHF283 (GFP-sirVDAC3), pHF284 (GFP-PACT) and pHF285 (GFP-Mps1-PACT). These plasmids were created as follows: to create the yeast two-hybrid bait plasmid pHF269, a PCR product corresponding to Mps1 amino acids 53–175 was cloned into pENTR-3C and transferred into pDest-32 using the Gateway® Recombination system (Invitrogen). In order to create 6his- and MBP-tagged constructs the VDAC1, VDAC2 and VDAC3 open reading frames were amplified by PCR from human MGC-verified cDNA clones obtained from Open Biosystems, cloned into the pENTR/D/SD/TOPO entry vector (Invitrogen) and transferred into pDEST17 and pKM596 using the Gateway® Recombination system (Invitrogen) to generate pHF270, pHF271, pHF272 and pHF273, respectively. In order to construct GST-tagged constructs pHF274, pHF275, pHF276 and pHF277, the corresponding DNA fragments were PCR-amplified from the entry clone and ligated into pGex6p-1. The vectors pMAL-c2X (New England Biolabs) and pGex6P-1 were used to express MBP and GST alone in bacteria. GFP-tagged mammalian expression constructs pHF278 (GFP-Mps153-175), pHF279 (GFP-VDAC3), pHF280 (VDAC3-GFP), pHF281 (GFP-VDAC3-PACT) and pHF282 (DsRed-VDAC3) were created using PCR to flank open-reading frames from the entry clones with KpnI and XbaI for cloning into pHF7, pHF148 and pHF252. The siRNA-resistant VDAC3 expression construct pHF283 (GFP-sirVDAC3) was created by site-directed mutagenesis of pHF276, using the QuikChange XL site-directed mutagenesis kit (Stratagene). pHF284 and pHF285 were created by inserting a 492 bp DNA fragment encoding the β globin intron downstream of the SV40 promoter in pHF148 (GFP-PACT) and pHF142 (GFP-Mps1-PACT). The sequences of PCR primers are available upon request. The identity of all constructs was verified by sequence analysis.
Cell culture
HEK293, HeLa S3, U2OS and NIH3T3 cells were cultured in DMEM and hTERT-RPE1 cells in DME/F-12 (1:1) (Hyclone) supplemented with 10% FBS (Atlanta Biologicals), 100 U/ml penicillin G and 100 μg/ml streptomycin (Hyclone) in the presence of 5% CO2. The expression of GFP-Mps1∆12/13 in HeLa GFP-Mps1∆12/13 cells14 that were maintained of 500 μg/ml G418 (Sigma) was induced by doxycycline (dox; Sigma) at 1 μg/ml. Proteasome-inhibition was achieved using treatment with 5 μM MG115 (Sigma) for 4 h. For BrdU incorporation, asynchronously growing cells were incubated with BrdU (40 μM; Sigma) for 4 h. S-phase arrest was achieved using a 24 h treatment with 4 mM hydroxyurea (HU; Sigma). For the centriole reduplication assay, the beginning of S-phase arrest was considered to be 24 h after the addition of HU, and centriole number was assessed 72 h after addition of HU.
DNA and siRNA transfections
Mammalian constructs were transfected using Effectene (Qiagen) in HeLa, Lipofectamine 2000 (Invitrogen) in HEK293 cells and FuGENE6 (Roche) in RPE1 cells. Stealth siRNAs directed against VDAC3 (siVDAC3–1: nucleotides 330–354; siVDAC3–2: nucleotides 625–649) and Mps1 (siMps1: nucleotides 1360–1384) obtained from Invitrogen and siGLO Lamin A/C siRNA (siControl) obtained from Dharmacon were used at a final concentration of 40 nM for transfection using Lipofectamine RNAiMAX (Invitrogen), and cells were analyzed 72 h after transfection.
RT-PCR
One μg of total RNA isolated using TRIzol reagent (Invitrogen) was used for cDNA synthesis using Superscript III first-strand synthesis system (Invitrogen), and the cDNA was further assayed using either conventional PCR or quantitative PCR with an Eco real-time PCR system (Illumina) using SYBR green-based chemistry (Applied Biosystems). Primers used were- VDAC1: 5′-TACAAGCGGGAGCACATT-3′ and 5′-CTGGGTCACTCGGGATTT-3′, VDAC2: 5′-TACCTTCTCACCAAACACAGG-3′ and 5′-AGCCCTCATAACCAAAGACA-3′, VDAC3: 5′-TGGACTTACCTTCACCCAGA-3′ and 5′-CTGCCAACACTAAAACAATCC-3′, GAPDH (endogenous control): 5′-AGGTCGGTGTGAACGGATTTG-3′ and 5′-TGTAGACCATGTAGTTGAGGTCA-3′, VDAC3 cDNA primer1- 5′-CCAAGTCTTGTAGTGGAGTGATG-3′, VDAC3 cDNA primer2- 5′-CCAAGTCTTGTAGTGGAGTG-3′, VDAC3 cDNA primer3- 5′-TCAGTGTAAGCATGACCAGA-3′. The specificity of the primer sets for respective VDAC genes and the absence of genomic DNA contamination in the cDNAs were verified by PCR.
Cytology
Antibodies and working dilutions for indirect immunofluorescence (IIF) were as follows: GTU-88 mouse anti-γ-tubulin, 1:200 (Sigma); rabbit anti-γ-tubulin, 1:200 (Sigma); affinity purified rabbit anti-Cetn2, 1:4,000,11 4G9 mouse anti-Mps1, 1:200 (H00007272-M02, Novus Biologicals); mouse anti-Sas6, 1:100 (Santa Cruz Biotechnology); rabbit anti-CP110, 1:500 (a kind gift from Dr. Brian Dynlacht, New York University School of Medicine); rabbit anti-Cep135, 1:500 (Abcam); rat anti-BrdU, 1:250 (Abcam); mouse anti-acetylated tubulin, 1:1,000 (Sigma); rabbit anti-VDAC3, 1:50 (Aviva Systems Biology or Sigma); rabbit anti-Ninein, 1:1000 (Abcam); mouse anti-Cep170, 1:500 (Invitrogen); mouse monoclonal anti-human mitochondria, 1:200 (Millipore), rabbit anti-GFP, 1:200 (Sigma). Secondary antibodies for IIF were goat or donkey anti-rabbit, donkey anti-mouse, or donkey anti-rat conjugated to Alexa 350 (1:200), Alexa 488 (1:1,000), Alexa 594 (1:1000), or Alexa 750 (1:200) (all from Invitrogen), and DNA was stained with Hoechst 33342 (Sigma). Cells were fixed with either PBS containing 4% formaldehyde (Ted Pella) and 0.2% Triton X-100 for 10 min at room temperature, or in methanol at –20°C for 10 min. For visualizing mitochondria, cells were incubated with 100 μM MitotrackerRed (Invitrogen) for 1 h prior to fixation in formaldehyde. For visualizing BrdU, cells fixed in methanol were stained with primary and secondary antibodies to cellular antigens, fixed again in methanol, treated with 2 N HCl for 30 min at room temperature, followed by staining with anti-BrdU antibody.46 The centrosomal level of Mps1 was measured as a ratio of the integrated signal of Mps1 in the vicinity of centrosomes to that of centrosomal γ-tubulin using a local background correction method based on our earlier quantitative IIF technique.10 Briefly, fixed cells on coverslips were immunostained for Mps1 and γ-tubulin and then imaged under identical conditions. After no neighbors deconvolution of the projection of image stacks along the Z-axis, background-corrected fluorescence intensity of the Mps1 (FMps1) and γ-tubulin (Fγ-tub) signals at the two closely spaced centrosomes of BrdU-positive cells were calculated from the integrated fluorescence intensities of each fluorophore in a small square box (FS) surrounding each centrosome, that of a larger square box (FL) surrounding the small box and the area of each square box (AS and AL), using the formula described by Howell et al.47; F = FS – {(FL – FS) × [AS/(AS – AS)]}. The ratio FMps1/Fγ-tub was determined for 25 cells for each sample. All images were acquired at ambient temperature using an Olympus IX-81 microscope, with a 63X or 100X Plan Apo oil immersion objective (1.4 numerical aperture) and a QCAM Retiga Exi FAST 1394 camera, and analyzed using the Slidebook software package (Intelligent Imaging Innovations).
Immunoprecipitation and immunoblotting
Cells were lysed in either HeLa lysis buffer (50 mM TRIS-HCl pH8.0, 150 mM NaCl and 1% NP-40) or RIPA buffer (50 mM TRIS-HCl pH8.0, 150 mM NaCl and 1% TritonX-100, 0.25% Na-Deoxycholate, 0.1% SDS). For immunoprecipitation experiments, 1 μg antibody [3E6 mouse anti GFP (Invitrogen) or C-19 rabbit anti-Mps1 (Santa Cruz Biotechnology)] coupled to Dynabeads ProteinG (Invitrogen) was incubated with the cellular extracts that had been pre-cleared by incubating with Dynabeads ProteinG alone (PC). After 4 h incubation with the cellular extracts, antibody-coupled beads were washed and the immunoprecipitated complex (IP) was extracted in SDS-PAGE sample buffer (under reducing condition unless specified) for immunoblotting. Since the visualization of VDAC3 bands in the immunoprecipitates of rabbit anti-Mps1 antibody were difficult in presence of IgG light chains and ProteinG molecules, the IP and PC were eluted in 100 mM Glycine-HCl, pH2.3 (to avoid the extraction of ProteinG) followed by precipitation using trichloroacetic acid (TCA) and re-solubilized in sample buffer without reducing agent (to avoid the dissociation of IgG chains). However, visualization of both VDAC3a and Mps1 required the reducing condition (addition of DTT), whereas the detection of VDAC3b was largely unaffected under non-reducing condition. Antibodies for immunoblot analysis were: 1:1,000 C-19 rabbit anti-Mps1 (Santa Cruz Biotechnology), 1:1,000 MDS rabbit anti-Mps1,101:2,000 rabbit anti-GFP (Sigma), 1:1,000 N1 mouse anti-Mps1 (Invitrogen), 1:20,000 DM1A mouse anti α-tubulin (Sigma), 1:1,000 rabbit anti-DsRed (Clonetech), 1:1,000 rabbit anti-VDAC3 (Aviva Systems Biology or Sigma), 1:20,000 mouse anti-GAPDH (Sigma), 1:1,000 rabbit anti-COX IV (Cell Signaling), 1:1,000 mouse anti-γ-tubulin (Sigma) and 1:500 rabbit anti-His (GenWay). Secondary antibodies (all used at 1:10,000 dilution) were Alexa680-conjugated donkey anti-mouse/rabbit (Invitrogen) and IRDye800-conjugated donkey anti-mouse/ rabbit (Rockland) for detection by Odyssey imaging system (LI-COR) or HRP-conjugated sheep anti-mouse IgG (GE Healthcare) for detection using SuperSignal West Femto Chemiluminescent Substrate (Thermo Scientific). The background-corrected intensities of the bands were calculated using Odyssey imaging system (LI-COR).
Cellular fractionation
HeLa or 293 cells were fractionated using Qproteome mitochondria isolation kit (Qiagen) according to the manufacturer’s instructions. Briefly, cells incubated in a lysis buffer were centrifuged at 1,000 × g for 10 min to produce a supernatant containing cytosolic proteins (Cy). The pellet was resuspended in disruption buffer using needle and re-centrifuged at 1,000 × g for 10 min to generate a pellet-containing nuclei, cell debris and unlysed cells (referred to as N/u). The supernatant was centrifuged at 6,000 × g for 10 min to pellet mitochondria (Mi) leaving the microsomal fraction in the supernatant (Mt). Both pellets were re-extracted in HeLa lysis buffer.
The sucrose density gradient fractionation to enrich the centrosomal proteins was performed according to previously described protocols.48,49 Briefly, asynchronous population of HeLa cells (5 × 107 cells) were treated with 1 μM nocodazol and 1 μg/ml cytochalasin D for 1 h, harvested and lysed in a buffer containing 1 mM Tris-Cl, pH 7.4, 0.4 mM DTT, 0.5 mM MgCl2, 0.5% NP-40 for 5 min. The lysate was centrifuged at 2,500 × g for 10 min to remove nuclei and cell debris. The supernatant was adjusted to 10 mM PIPES (pH 7.2), treated with DNase I (2 units/mL) for for 30 min and then underlaid with a 60% sucrose solution in 10 mM PIPES, 0.4 mM DTT and 0.1% Triton X-100 to sediment the centrosomes into the sucrose cushion by centrifuging at 10,000 × g for 30 min. This crude centrosome preparation (~6 ml) was further fractionated by discontinuous sucrose gradient (2.25 mL of 70%, 1.35 mL of 50% and 1.35 mL of 40% sucrose in 10 mM PIPES, 0.4 mM DTT and 0.1% Triton X-100) ultracentrifugation at 120,000 × g for 1 h at 4oC. Fractions (500 μl) were collected from the bottom and supplemented with 10% glycerol. A protease inhibitor solution was present at all steps of both the fractionation that were performed on ice or at 4°C.
Protein expression and purification from bacteria
All fusion proteins were expressed in E. coli BL21 (DE3). For protein purification, cells carrying GST- or MBP-fusion constructs were induced at 18°C with 250 μM IPTG for 16 h, harvested and were lysed in either GST-lysis buffer (20 mM Tris-Cl, pH 7.4, 10 mM NaCl, 2 mM MgCl2 supplemented with 2 mM DTT, 1% TritonX-100, 1 mM PMSF) or MBP-lysis buffer (20 mM Tris-Cl, pH 7.4, 200 mM NaCl, 1 mM EDTA supplemented with 1 mM DTT, 1 mM PMSF) followed by centrifugation at 15,000 × g for 30 min and purification by respective affinity chromatography. After extensive washing with a buffer containing 200 mM NaCl, purified protein bound-beads were stored in storage buffer (50 mM Tris-Cl, pH 7.4, 25% glycerol, 5 mM MgCl2) at -80°C.
Pull-down assays
Cell lysates were pre-cleared on the corresponding beads (either glutathione Sepharose or amylose) before being incubated with equilibrated bait protein-bound beads, for 4 h at 4°C. After washing the beads using HeLa lysis buffer, the proteins bound with the beads were extracted with 2X SDS-PAGE sample buffer.
Yeast two-hybrid assay
pHF282 was transformed into the Saccharomyces cerevisiae reporter strain Mav203 and the pre-made human cDNA library in pPC86 (Invitrogen) was screened against the CLD as instructed by the ProQuestTM two-hybrid system with GatewayTM Technology (Invitrogen). Over 1,000,000 transformants were screened to obtain 1,800 colonies that grew in the absence of His, Leu and Trp. Further screening in presence of 25 mM 3-Amino-1,2,4-triazole (3-AT) resulted 120 colonies, 62 of which also grew in the absence of uracil. Finally, four of these transformants were strongly positive for the lacZ reporter in a β-galactosidase activity assay. Plasmids from these positive clones were extracted and sequenced to identify the corresponding gene using BLAST in the NCBI database.
Protein alignments
Vertebrate Mps1 sequences were obtained from GenBank using NCBI (www.ncbi.nlm.nih.gov/protein). Regions corresponding to amino acids 52–180 were aligned using ClustalW (www.ebi.ac.uk/Tools/msa/clustalw2/) and color coded using BioEdit software (Ibis Biosciences).
Cell viability assay
1X104 RPE1 cells growing in a 96-well plate were treated with either Erastin or for 16 h or FCCP for 4 h at the indicated final concentration. DMSO was used as the solvent control. Cell viability was determined on triplicate samples of each treatment group using the CellTiter-Glo luminescent cell viability assay (Promega), a luminescent assay for measuring cellular ATP.
Supplementary Material
Acknowledgments
We are grateful to Drs. Daniel Schoenberg and Chandrama Mukherjee of The Ohio State University for their help in real-time PCR and Dr. Thomas Giddings of University of Colorado for Immuno-EM. This work was supported by a National Institutes of Health grant (GM77311) and an Ohio Cancer Research Associates seed grant to H.A.F., and an Up on the Roof fellowship from The Ohio State University Comprehensive Cancer Center to S.M.
Glossary
Abbreviations:
- CLD
centrosome localization domain
- dox
doxycycline
- DsRed
monomeric Discosoma sp. red fluorescent protein
- FCCP
carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone
- siCon
control siRNA
- siVDAC3
VDAC3-siRNA
- siMps1
Mps1-siRNA
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21927
References
- 1.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–82. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol. 2000;149:317–30. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azimzadeh J, Marshall WF. Building the centriole. Curr Biol. 2010;20:R816–25. doi: 10.1016/j.cub.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leidel S, Delattre M, Cerutti L, Baumer K, Gönczy P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat Cell Biol. 2005;7:115–25. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- 5.Nakazawa Y, Hiraki M, Kamiya R, Hirono M. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr Biol. 2007;17:2169–74. doi: 10.1016/j.cub.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 6.Pike AN, Fisk HA. Centriole assembly and the role of Mps1: defensible or dispensable? Cell Div. 2011;6:9. doi: 10.1186/1747-1028-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–78. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 8.Kang J, Chen Y, Zhao Y, Yu H. Autophosphorylation-dependent activation of human Mps1 is required for the spindle checkpoint. Proc Natl Acad Sci USA. 2007;104:20232–7. doi: 10.1073/pnas.0710519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisk HA, Mattison CP, Winey M. Human Mps1 protein kinase is required for centrosome duplication and normal mitotic progression. Proc Natl Acad Sci USA. 2003;100:14875–80. doi: 10.1073/pnas.2434156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasbek C, Yang CH, Fisk HA. Antizyme restrains centrosome amplification by regulating the accumulation of Mps1 at centrosomes. Mol Biol Cell. 2010;21:3878–89. doi: 10.1091/mbc.E10-04-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang CH, Kasbek C, Majumder S, Yusof AM, Fisk HA. Mps1 phosphorylation sites regulate the function of centrin 2 in centriole assembly. Mol Biol Cell. 2010;21:4361–72. doi: 10.1091/mbc.E10-04-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasbek C, Yang CH, Yusof AM, Chapman HM, Winey M, Fisk HA. Preventing the degradation of mps1 at centrosomes is sufficient to cause centrosome reduplication in human cells. Mol Biol Cell. 2007;18:4457–69. doi: 10.1091/mbc.E07-03-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisk HA, Winey M. The mouse Mps1p-like kinase regulates centrosome duplication. Cell. 2001;106:95–104. doi: 10.1016/S0092-8674(01)00411-1. [DOI] [PubMed] [Google Scholar]
- 14.Kasbek C, Yang CH, Fisk HA. Mps1 as a link between centrosomes and genomic instability. Environ Mol Mutagen. 2009;50:654–65. doi: 10.1002/em.20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Cheng X, Zhang Y, Li S, Cui H, Zhang L, et al. Phosphorylation of Mps1 by BRAF(V600E) prevents Mps1 degradation and contributes to chromosome instability in melanoma. Oncogene. 2012 doi: 10.1038/onc.2012.94. [DOI] [PubMed] [Google Scholar]
- 16.Lingle WL, Barrett SL, Negron VC, D’Assoro AB, Boeneman K, Liu W, et al. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci USA. 2002;99:1978–83. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sampson MJ, Lovell RS, Craigen WJ. The murine voltage-dependent anion channel gene family. Conserved structure and function. J Biol Chem. 1997;272:18966–73. doi: 10.1074/jbc.272.30.18966. [DOI] [PubMed] [Google Scholar]
- 18.Rahmani Z, Maunoury C, Siddiqui A. Isolation of a novel human voltage-dependent anion channel gene. Eur J Hum Genet. 1998;6:337–40. doi: 10.1038/sj.ejhg.5200198. [DOI] [PubMed] [Google Scholar]
- 19.Hinsch KD, De Pinto V, Aires VA, Schneider X, Messina A, Hinsch E. Voltage-dependent anion-selective channels VDAC2 and VDAC3 are abundant proteins in bovine outer dense fibers, a cytoskeletal component of the sperm flagellum. J Biol Chem. 2004;279:15281–8. doi: 10.1074/jbc.M313433200. [DOI] [PubMed] [Google Scholar]
- 20.Nakagawa Y, Yamane Y, Okanoue T, Tsukita S, Tsukita S. Outer dense fiber 2 is a widespread centrosome scaffold component preferentially associated with mother centrioles: its identification from isolated centrosomes. Mol Biol Cell. 2001;12:1687–97. doi: 10.1091/mbc.12.6.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu ST, Chan GK, Hittle JC, Fujii G, Lees E, Yen TJ. Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol Biol Cell. 2003;14:1638–51. doi: 10.1091/mbc.02-05-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S, Thebault P, Freschi L, Beaufils S, Blundell TL, Landry CR, et al. Characterization of spindle checkpoint kinase Mps1 reveals domain with functional and structural similarities to tetratricopeptide repeat motifs of Bub1 and BubR1 checkpoint kinases. J Biol Chem. 2012;287:5988–6001. doi: 10.1074/jbc.M111.307355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shoshan-Barmatz V, De Pinto V, Zweckstetter M, Raviv Z, Keinan N, Arbel N. VDAC, a multi-functional mitochondrial protein regulating cell life and death. Mol Aspects Med. 2010;31:227–85. doi: 10.1016/j.mam.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 24.De Pinto V, Guarino F, Guarnera A, Messina A, Reina S, Tomasello FM, et al. Characterization of human VDAC isoforms: a peculiar function for VDAC3? Biochim Biophys Acta. 2010;1797:1268–75. doi: 10.1016/j.bbabio.2010.01.031. [DOI] [PubMed] [Google Scholar]
- 25.Müller H, Schmidt D, Steinbrink S, Mirgorodskaya E, Lehmann V, Habermann K, et al. Proteomic and functional analysis of the mitotic Drosophila centrosome. EMBO J. 2010;29:3344–57. doi: 10.1038/emboj.2010.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sampson MJ, Ross L, Decker WK, Craigen WJ. A novel isoform of the mitochondrial outer membrane protein VDAC3 via alternative splicing of a 3-base exon. Functional characteristics and subcellular localization. J Biol Chem. 1998;273:30482–6. doi: 10.1074/jbc.273.46.30482. [DOI] [PubMed] [Google Scholar]
- 27.Maro B, Marty MC, Bornens M. In vivo and in vitro effects of the mitochondrial uncoupler FCCP on microtubules. EMBO J. 1982;1:1347–52. doi: 10.1002/j.1460-2075.1982.tb01321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guarguaglini G, Duncan PI, Stierhof YD, Holmström T, Duensing S, Nigg EA. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell. 2005;16:1095–107. doi: 10.1091/mbc.E04-10-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Azimzadeh J, Hergert P, Delouvée A, Euteneuer U, Formstecher E, Khodjakov A, et al. hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J Cell Biol. 2009;185:101–14. doi: 10.1083/jcb.200808082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. Cep152 interacts with Plk4 and is required for centriole duplication. J Cell Biol. 2010;191:721–9. doi: 10.1083/jcb.201006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salisbury JL, Suino KM, Busby R, Springett M. Centrin-2 is required for centriole duplication in mammalian cells. Curr Biol. 2002;12:1287–92. doi: 10.1016/S0960-9822(02)01019-9. [DOI] [PubMed] [Google Scholar]
- 32.Yagoda N, von Rechenberg M, Zaganjor E, Bauer AJ, Yang WS, Fridman DJ, et al. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447:864–8. doi: 10.1038/nature05859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang WS, Stockwell BR. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem Biol. 2008;15:234–45. doi: 10.1016/j.chembiol.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gillingham AK, Munro S. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 2000;1:524–9. doi: 10.1093/embo-reports/kvd105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sang L, Miller JJ, Corbit KC, Giles RH, Brauer MJ, Otto EA, et al. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 2011;145:513–28. doi: 10.1016/j.cell.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams CL, Li C, Kida K, Inglis PN, Mohan S, Semenec L, et al. MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J Cell Biol. 2011;192:1023–41. doi: 10.1083/jcb.201012116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cerqua C, Anesti V, Pyakurel A, Liu D, Naon D, Wiche G, et al. Trichoplein/mitostatin regulates endoplasmic reticulum-mitochondria juxtaposition. EMBO Rep. 2010;11:854–60. doi: 10.1038/embor.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ibi M, Zou P, Inoko A, Shiromizu T, Matsuyama M, Hayashi Y, et al. Trichoplein controls microtubule anchoring at the centrosome by binding to Odf2 and ninein. J Cell Sci. 2011;124:857–64. doi: 10.1242/jcs.075705. [DOI] [PubMed] [Google Scholar]
- 39.Kanai M, Ma Z, Izumi H, Kim SH, Mattison CP, Winey M, et al. Physical and functional interaction between mortalin and Mps1 kinase. Genes Cells. 2007;12:797–810. doi: 10.1111/j.1365-2443.2007.01091.x. [DOI] [PubMed] [Google Scholar]
- 40.Xu X, Colombini M. Autodirected insertion: preinserted VDAC channels greatly shorten the delay to the insertion of new channels. Biophys J. 1997;72:2129–36. doi: 10.1016/S0006-3495(97)78855-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ujwal R, Cascio D, Colletier JP, Faham S, Zhang J, Toro L, et al. The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating. Proc Natl Acad Sci USA. 2008;105:17742–7. doi: 10.1073/pnas.0809634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krantz DD, Zidovetzki R, Kagan BL, Zipursky SL. Amphipathic beta structure of a leucine-rich repeat peptide. J Biol Chem. 1991;266:16801–7. [PubMed] [Google Scholar]
- 43.Lai PY, Wang CY, Chen WY, Kao YH, Tsai HM, Tachibana T, et al. Steroidogenic Factor 1 (NR5A1) resides in centrosomes and maintains genomic stability by controlling centrosome homeostasis. Cell Death Differ. 2011;18:1836–44. doi: 10.1038/cdd.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Decker WK, Craigen WJ. The tissue-specific, alternatively spliced single ATG exon of the type 3 voltage-dependent anion channel gene does not create a truncated protein isoform in vivo. Mol Genet Metab. 2000;70:69–74. doi: 10.1006/mgme.2000.2987. [DOI] [PubMed] [Google Scholar]
- 45.Uetake Y, Loncarek J, Nordberg JJ, English CN, La Terra S, Khodjakov A, et al. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J Cell Biol. 2007;176:173–82. doi: 10.1083/jcb.200607073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsou MF, Wang WJ, George KA, Uryu K, Stearns T, Jallepalli PV. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell. 2009;17:344–54. doi: 10.1016/j.devcel.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howell BJ, Hoffman DB, Fang G, Murray AW, Salmon ED. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J Cell Biol. 2000;150:1233–50. doi: 10.1083/jcb.150.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moudjou M. Isolation of Centrosomes from Cultured Animal cells. Cell Biology: A Laboratory Handbook. 1994:595–604. [Google Scholar]
- 49.Puklowski A, Homsi Y, Keller D, May M, Chauhan S, Kossatz U, et al. The SCF-FBXW5 E3-ubiquitin ligase is regulated by PLK4 and targets HsSAS-6 to control centrosome duplication. Nat Cell Biol. 2011;13:1004–9. doi: 10.1038/ncb2282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.