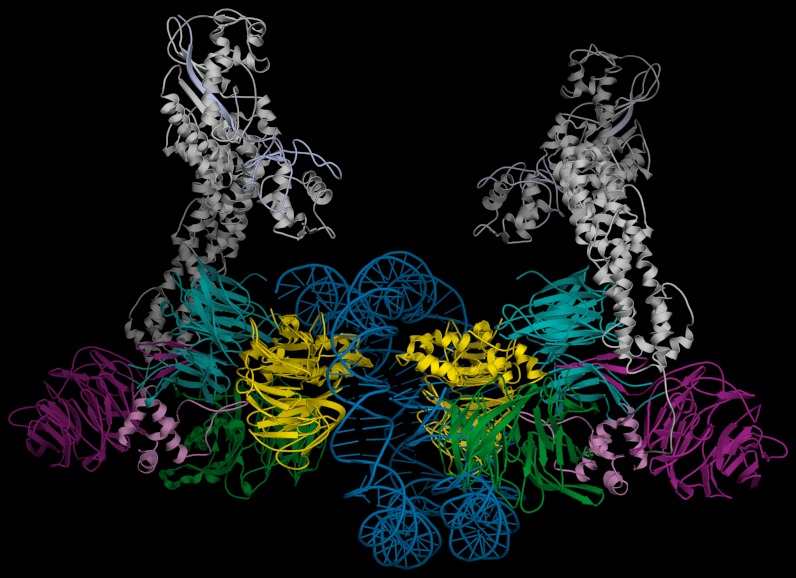
Fig. P1.
Composite model of a dimeric DDB1-CUL4ADDB2 ubiquitin ligase-nucleosome complex. A model of a dimeric DDB1-CUL4ADDB2 in a complex with a nucleosome core particle, generated according to the relative subunit organization found in the dimeric UV-DDB-AP24 crystal structure. The intermolecular orientation of the CUL4A-Rbx (in gray) subunit resulted from superimposing two copies of the respective BPA and BPC domains of the DDB1 subunit in the DDB1-CUL4A-Rbx structure (accession number 2HYE) onto the corresponding DDB1 domains (BPA in blue, BPC in green, BPB in purple) in the dimeric UV-DDB crystal structure. The nucleosome core particle (PDB ID code 1AOI) was aligned by superpositioning the respective sugar-phosphate backbones of the nucleosomal DNA (in blue) onto the backbones of two adjacent AP24 oligodeoxynucleotides (not shown). Remarkably, the available molecular volume between two AP24 DNA molecules accommodates the spatial requirements of the nucleosome particle, with minor adjustments of the two DDB2 subunits (in yellow). For visual clarity, the histones of the nucleosome core particle and the AP24 subunits are not depicted in this figure. For a view showing the superpositioning of the nucleosome onto the two AP24 oligodeoxynucleotide molecules of the UV-DDB complex, refer to Fig. 4 of the complete on-line publication.