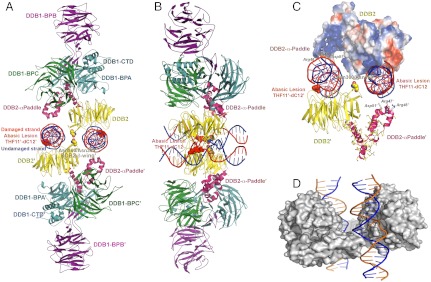
Fig. 2.
Structure of the dimeric human UV-DDB in a complex with damaged DNA. (A) The dimeric UV-DDB subunit organization, shown in ribbon depiction, with each domain colored and labeled accordingly: yellow, DDB2 β-propeller; red, DDB2 N-terminal-α paddle; blue, DDB1 BPA; green, DDB1 BPC; and purple, DDB1 BPB. The 24-bp oligodeoxynucleotide (AP24) contains an abasic lesion site (THF11), with the phosphor-deoxyribose backbone of the damaged strand colored in red and the undamaged strand colored in blue. Each DDB2 subunit is bound to an AP24 oligonucleotide, with DDB2 residues Asn360/Asn360’ straddling the twofold symmetry axis, forming H bonds across the dimer interface. The surface of the Asn360/Asn360’ pair (colored using standard atom convention) is located in a loop spanning two antiparallel β-strands (β-wing). The abasic lesion site in AP24 is marked by the surface mesh drawn around nucleotides THF11/dC12 in their flipped, extra-helical configuration. The β-wing is sandwiched between the two AP24 oligodeoxynucleotides, astride of the twofold axis of rotation relating the monomer subunits in the dimeric DDB2. Residues on the leading β-strand and loop form contacts with the undamaged DNA strand whereas residues on the loop and the retreating β-strand form contacts with the neighboring undamaged DNA strand. Both sets of contacts are predominantly electrostatic in nature, thus largely sequence independent. (B) Same as A but rotated 90 degrees and tilted slightly to show both DNA molecules. (C) Electrostatic potential surfaces of the DDB2 N-terminal domain complement the charge characteristics of the DDB1 BPC domain and the DNA phosphor-deoxyribose interfaces, resulting in favorable electrostatic neutralization. Contacts between residues on the β-wing region form contacts with the DNA bound at its immediate active site and with the neighbouring DNA molecule bound to the second monomer of DDB2 in the dimer. Extensive interactions between residues on the N-terminal-helical domain (α-paddle) and the neighboring DNA molecule augment the intermolecular associations, contributing to the high affinity of damaged DNA binding. (D) The skewed positioning of the DNA binding surface can now be understood in terms of the DDB2 dimer interface, located adjacent to the DNA binding site, at a loop bridging blades 6 and 7 of the β-propeller (β-wing) of DDB2. To accommodate the steric constraints imposed through dimerization along with DNA binding, the two adjacent sites are positioned diametrically across one face of the molecular surface of DDB2, readily seen in the dimeric DDB2-DNA (AP24) complex.