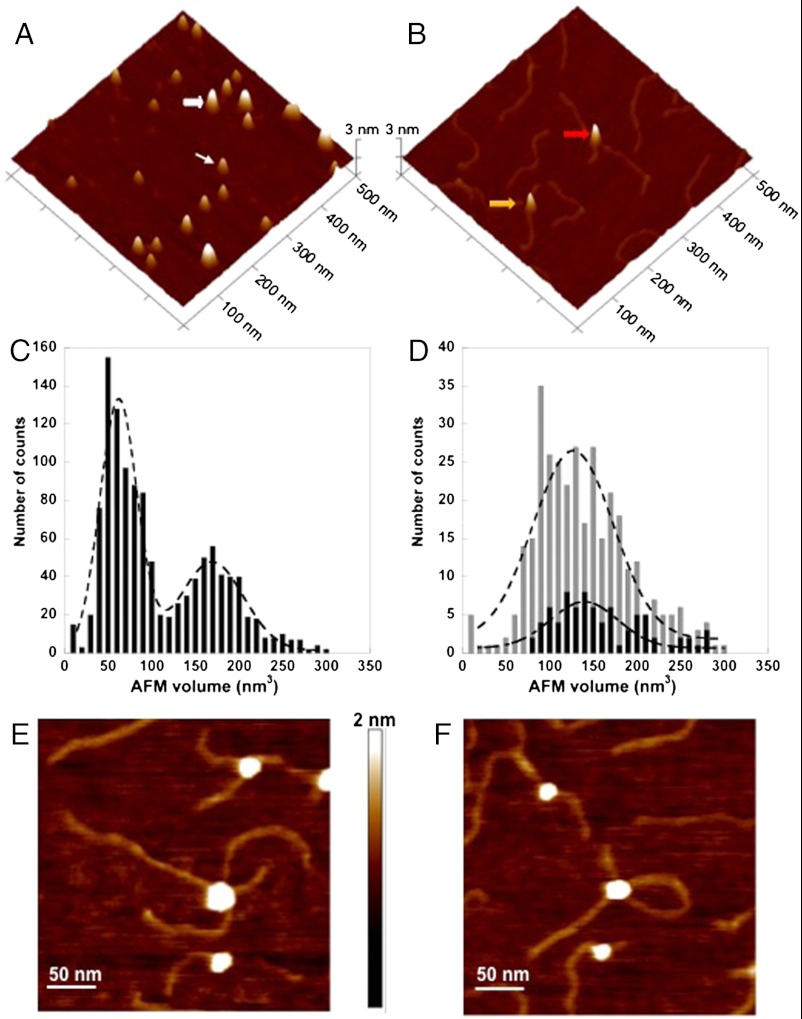
Fig. 3.
AFM imaging shows that damaged DNA binding promotes the dimerization of the DDB1-DDB2 heterodimer. (A) A representative surface plot of UV-DDB (50 nM) in the absence of DNA. The thin and wide white arrows point to molecules consistent with the size of the UV-DDB monomer (DDB1-DDB2 heterodimer) and trimer of UV-DDB, respectively. (B) Representative surface plot of UV-DDB (50 nM) in the presence of UV-irradiated 517 bp PCR fragments (25 nM). The yellow and red arrows point to dimeric UV-DDB [(DDB1-DDB2)2] binding to one and two molecules of duplex DNA, respectively. (C) AFM volume analysis of free UV-DDB (n = 1,160). (D) AFM volume analysis of UV-DDB on one strand (gray bars, n = 339) and two strands (black bars, n = 79) of duplex DNA. The images in A and B are at 500 nm × 500 nm and 3 nm in height. (Bottom) The dashed lines (C, free in solution, and D, bound to DNA) represent Gaussian fits to the data. Field view images of UV-DDB binding to separate DNA molecules (E) or two different regions of the same DNA molecule (F). The images are at 300 nm × 300 nm and 2 nm in height.