Fig. 3.
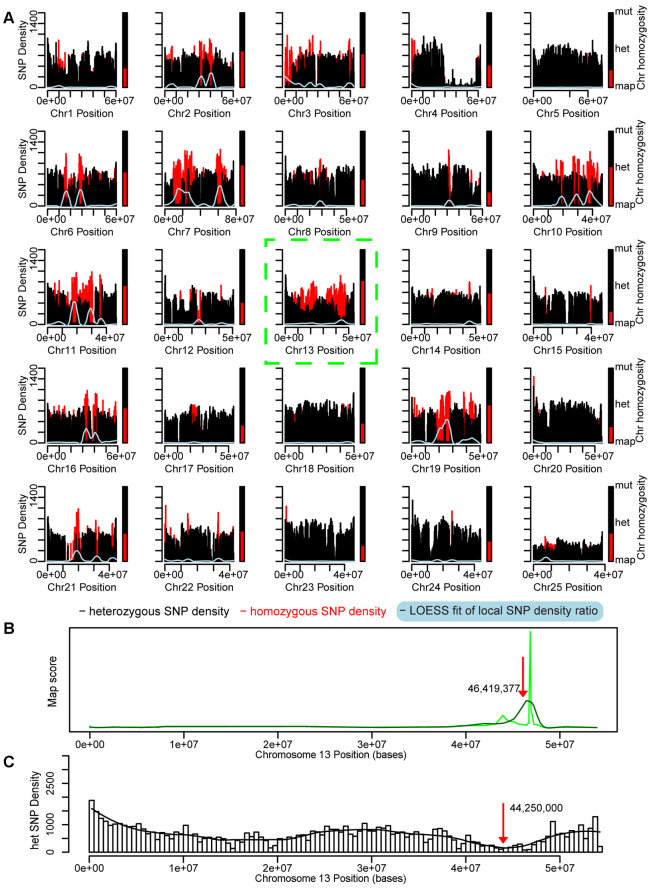
Homozygosity mapping and filtering of an158-3/cdh23nl9. (A) Chromosomal SNP density and homozygosity map of an158-3/cdh23nl9. Red bars are homozygous SNP counts, black bars are heterozygous SNP counts. Homozygous-to-heterozygous ratios were fitted for trend recognition using local regression (LOESS; light blue). Bars next to individual chromosomes show the degree of homozygosity (red bar, vs heterozygosity in black). Chromosome 13 shows the highest degree of homozygosity and thus probably harbors the mutation. (B) Scan of chromosome 13 for a peak in local homozygosity and fit peaks at 46,419,377 (red arrow). (C) A SNP density histogram of all detected heterozygous SNPs on chromosome 13. Loss of heterozygosity is calculated by fitting to the density histogram (black line). The minimum of the fit is indicated (red arrow). The average of the resulting values from the graphs shown in B and C was used to position the center of the critical interval. Variant effect prediction of all detected homozygous SNPs within the critical interval after filtering reveals one candidate SNP. SNP 13_43745899_G/A is located in the essential part of a splice acceptor site for ‘exon 7’ (ENSDARE00000862400) of cdh23, leading to a frameshift by mis-splicing and truncation within the extracellular domain (Table 3).