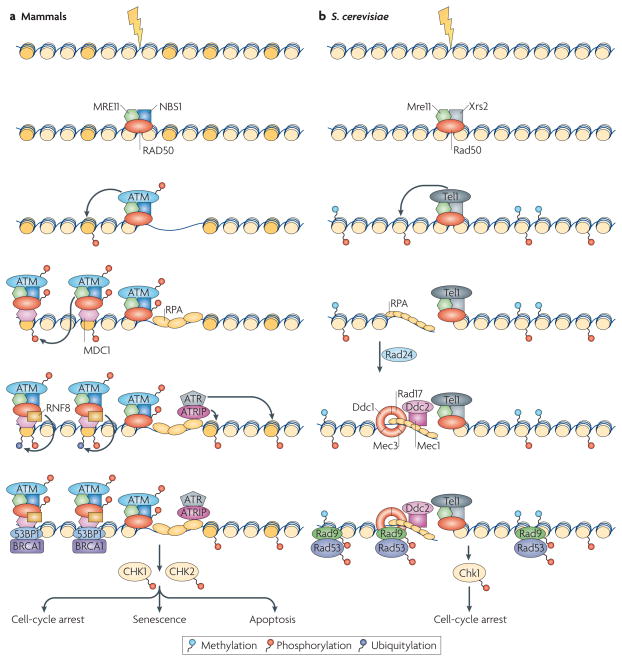
Figure 1. Assembly of DNA-damage response complexes.
a | In mammalian cells, the presence of a double-stranded DNA break (DSB) is originally sensed by the MRN (MRE11–RAD50–NBS1) sensor complex. The recruitment of MRN activates the transducer kinase ATM, which associates with DSBs and phosphorylates the histone variant H2AX (dark yellow) in DSB-flanking nucleosomes. The mediator MDC1 protein then binds to γ-H2AX and recruits additional copies of MRN and ATM, leading to spreading of the repair machinery along the chromosome. MDC1 also recruits ubiquitin ligase activities (for example, RNF8) that are responsible for the recruitment of downstream factors such as 53BP1 and BRAC1. DNA is then resected to single-stranded DNA (ssDNA) and recognized by replication protein A (RPA), which results in the recruitment of ATR through its interacting partner ATRIP. Both the ATM- and the ATR-dependent branches of the pathway, independently or in concert, lead to the activation of the checkpoint kinases CHK1 and CHK2. b | In Saccharomyces cerevisae, the DNA-damage response (DDR) cascade starts with the binding of the MRX (Mre11–Rad50–Xrs2) complex to the DSB. MRX recruits the Tel1 kinase, which phosphorylates histone H2A. After resection, ssDNA is sensed by RPA, which recruits the Ddc1–Mec3–Rad17 (known as 9–1–1 complex in mammals) sensor complex (Rad24 acts as a clamp loader) and the Ddc2–Mec1 complex independently. Next, Rad9 is recruited through its affinity for phosphorylated H2A and methylated H3, and the checkpoint kinases Chk1 and Rad53 are activated.