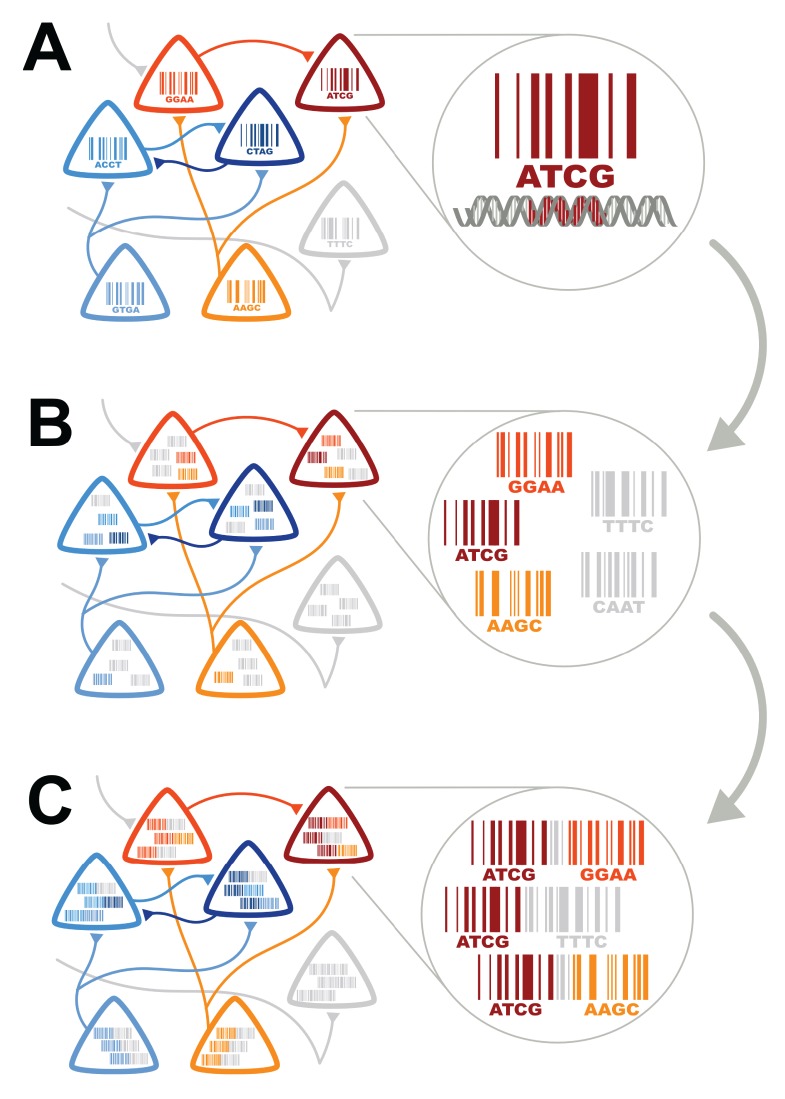
Figure 2. Converting connectivity into a sequencing problem can be broken down conceptually into three components.
Each component of BOINC has many possible implementations. (A) First, each neuron must be labeled with a unique sequence of nucleotides—a DNA “barcode”. (B) Second, barcodes from synaptically connected neurons must be associated with one another, so that each neuron can be thought of as a “bag of barcodes”: copies of its own “host” barcode and copies of “invader” barcodes from synaptic partners. (C) Finally, host and invader barcodes must be joined into barcode pairs. These pairs can be subjected to high-throughput sequencing.