Abstract
Background
The objective of the study was to investigate the role of genes (HSD3B1, CYP17A1, CYP19A1, HSD17B2, HSD17B1) involved in the steroid hormone biosynthesis pathway and progesterone receptor (PGR) in the etiology of gastric cancer in a population-based two-phase genetic association study.
Methods
In the discovery phase, 108 candidate SNPs in the steroid hormone biosynthesis pathway related genes and PGR were analyzed in 76 gastric cancer cases and 322 controls in the Korean Multi-Center Cancer Cohort. Statistically significant SNPs identified in the discovery phase were re-evaluated in an extended set of 386 cases and 348 controls. Pooled- and meta-analyses were conducted to summarize the results.
Results
Of the 108 SNPs in steroid hormone biosynthesis pathway related genes and PGR analyzed in the discovery phase, 23 SNPs in PGR in the recessive model and 10 SNPs in CYP19A1 in the recessive or additive models were significantly associated with increased gastric cancer risk (p<0.05). The minor allele frequencies of the SNPs in both the discovery and extension phases were not statistically different. Pooled- and meta-analyses showed CYP19A1 rs1004982, rs16964228, and rs1902580 had an increased risk for gastric cancer (pooled OR [95% CI] = 1.22 [1.01–1.48], 1.31 [1.03–1.66], 3.03 [1.12–8.18], respectively). In contrast, all PGR SNPs were not statistically significantly associated with gastric cancer risk.
Conclusions
Our findings suggest CYP19A1 that codes aromatase may play an important role in the association of gastric cancer risk and be a genetic marker for gastric cancer susceptibility.
Introduction
Gastric cancer mortality is the second greatest in the world [1]. Gastric cancer incidence is approximately two times greater among men than women in many regions of the world [2], and the ratio becomes smaller after 60 years of age when most women reach menopause. Gastric cancer incidence in men is more than double than in women in the Korean population (62.2 vs. 24.6 per 100,000 persons) [3]. This global consistency of a high male to female incidence ratio in gastric cancer may be due to a hormonal difference between men and women. Thus, it has been hypothesized that female sex steroid hormones, estrogen and progesterone, may play a protective role in gastric cancer incidence.
Although inconsistent, epidemiological studies support this hypothesis. Many epidemiologic studies reported a decreased risk for gastric cancer with greater lifetime exposure to endogenous estrogen [4]–[11], whereas some studies showed no association [12]–[16]. Animal and in vitro studies also support this hypothesis. Female and castrated rats had a lower incidence of gastric cancer than non-treated male rats in N-methyl-N’-nitro-N-nitrosoguanidine carcinogenesis model [17]. In H. pylori-induced gastric cancer mouse model, 17 beta-estradiol acted as a protective factor in gastric carcinogenesis [18]. Estrogen demonstrated an increase in apoptosis in AGS human gastric cancer cells [19]. In addition, estrogen stimulated expression of trefoil peptides that are important in mucosal protection in the stomach [20]. Although published studies on estrogen influence on gastric cancer risk are inconsistent, a recent meta-analysis supports longer exposure to estrogen effects of either ovarian or exogenous origin may decrease gastric cancer risk [21].
Estrogen and progesterone are synthesized in the steroid hormone biosynthesis pathway. Steroid hormone receptors such as estrogen and progesterone have been identified and are expressed in gastric mucosa and cancer tissues [22]–[26]. Therefore, steroid hormone biosynthesis pathway and their receptors can be altered by genetic variations of related genes, thereby altering and contributing to individual susceptibility to gastric cancer. Of hormonal receptors, in particular, we focused on the progesterone receptor (PGR) because progesterone might be a major contributor for gastric carcinogenesis than estrogen. An animal study [27] showed that onapristone, a progesterone anatagonist, inhibited gastric tumor growth as well as estradiol-stimulated growth.
The hypothesis of the current study is genetic polymorphisms involved in the steroid hormone biosynthesis pathway and PGR can influence individual susceptibility in the development of gastric cancer. To investigate the hypothesis, a two-phase genetic association study was conducted: 1) the discovery phase was a candidate gene approach analysis focusing on five genes involved in the steroid biosynthesis pathway (HSD3B1, CYP17A1, CYP19A1, HSD17B2, and HSD17B1) and the hormone receptor gene (PGR); 2) the extension phase further examined the most significant SNPs identified in the discovery analysis.
Materials and Methods
Study Population
In the discovery phase, the population-based nested case-control study population was recruited from the Korean Multi-Center Cancer Cohort (KMCC), a community-based prospective cohort of participants recruited from four urban and rural areas in Korea (Haman, Chungju, Uljin, and Youngil) from 1993 through 2004 [28]. Participants completed detailed standardized interview based questionnaires on general lifestyle, medical history, physical activity, diet, reproductive factors, pesticide exposure, and additional environmental factors. Blood and spot urine samples were collected and stored at −70°C and −20°C, respectively.
In December 31, 2002, 136 gastric cancer cases in the KMCC were identified through computerized record linkages to the national cancer registry, the national death certificate, and the health insurance medical records. The passive follow-up methods were reported to be 99% efficient and completeness was assured [29]. Cases diagnosed before recruitment (N = 36) and without blood samples (N = 16) were excluded. Cancer-free controls were randomly selected from the KMCC population. There were four controls matched to each gastric cancer case by incidence density sampling based on age (±5 years), sex, residential district, and enrollment. Additionally, eight cases and 14 controls were excluded due to insufficient DNA or poor genotyping. Finally, 76 cases and 322 controls were included in the discovery phase.
In the extension phase, 388 gastric cancer case-control sets were selected as follows. There were 95 new gastric cancer cases and 52 prevalent cases in December 2008 and 52 additional cases whose blood samples were later obtained from the KMCC. In addition, from March 2002 to September 2006, 490 newly diagnosed gastric cancer patients from two university hospitals in Korea that were Chungnam University Hospital and Hanyang University GURI Hospital were identified. Epidemiological data and venous blood samples were collected at time of diagnosis or prior to gastric cancer surgery. Among them, 189 cases with sufficient DNA samples and informed consent were included. Community-based controls matched by age (±5 years), sex, and enrollment year from 2001 to 2005 were randomly selected from the KMCC. There were two cases and 40 controls excluded due to poor genotyping and insufficient sample. Finally, 386 cases and 348 controls were included in the extension phase. Pooled and meta-analyses included 462 cases and 670 controls.
Ethics Statement
All participants provided written informed consent before entering the studies. The study protocols for the KMCC and current nested case-control studies were approved by the institutional review boards of Seoul National University Hospital and the National Cancer Center of Korea (H-0110-084-002, C-0907-044-2861-170), and Hanyang University Hospital (2003–4).
Candidate Gene and SNP Selection
There were seven genes in the discovery phase selected from the literature review that were as follows: progesterone receptor (PGR); cytochrome P450, family 19, subfamily A, polypeptide 1 (CYP19A1); cytochrome P450, family 17, subfamily A, polypeptide 1 (CYP17A1); hydroxysteroid (17-beta) dehydrogenase 1 (HSD17B1); hydroxysteroid (17-beta) dehydrogenase 2 (HSD17B2); hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 (HSD3B1).
Candidate single nucleotide polymorphisms (SNP) from selected genes were selected according to the following criteria: 1) reported to have a possible functional significance in previous studies; 2) minor allele frequency (MAF)>0.05 in Asian databases such as SNP500Cancer, HapMap or CGAP using dbSNP IDs (http://www.ncbi.nlm.nih.gov/SNP); 3) concurrently, MAF>0.05 in HapMap Japanese (JET). Finally, 117 SNPS with a design score = 1.1, r2>0.8 in five candidate genes in the steroid hormone biosynthesis pathway and PGR were genotyped. There were 105 SNPs located in the intron region, eight SNPs located in the promoter region (flanking region or UTR), and four SNPs located in the coding region (Appendix S1).
In the extension phase, SNPs were selected as follows. For PGR, in the discovery analysis, 23 SNPs were significant and created one large Haploblock. There were two SNPs of the 23 SNPs located in the coding or 3UTR region. The raw and permutated p-values were less than 0.04. For CYP19A1, there were ten significant SNPs (raw p-value<0.05) located in the intron region. CYP19A1 created six blocks and the significant SNPs in the discovery phase were located in Blocks 4, 5, and 6.
Genotyping
Genomic DNA concentrations were measured for all study subjects by a spectrophotometer (NanoDrop ND-1000, NanoDrop Technologies). Genotyping in the discovery phase was performed using GoldenGateTM assay (Illumina®, San Diego, CA). Of the 117 SNPs, nine SNPs were deemed unusable due to failure of genotyping (rs6203 andrs9939740), SNP call rate<90% (rs2236780, rs12594293, rs12592697, rs597255, and rs2830), monomorphism (rs7175531, rs4243229), and were excluded in the analysis. Finally, we analyzed 108 SNPs in six genes (genotyping rate of 99.5%) in 76 cases and 322 controls. To ensure quality control and evaluate intra-subject concordance rate, 52 duplicate samples were randomly distributed in the genotyping plate. Concordance rates for all assays were greater than 99%.
Genotyping in the extension phase was performed using the IlluminaVeraCodeGoldenGate Assay with BeadXpress according to the manufacturer’s protocol (Illumina, San Diego, CA, USA) [16]. To ensure the reliability of the two different genotyping methods, 135 samples (59 cases and 76 controls) were genotyped by both the Genome-Wide Human SNP Array 5.0 and the IlluminaVeraCodeGoldenGate Assay, and the concordance rate was >98.2%. Because of the high concordance rate, all samples were included in the analysis; discordant samples were not eliminated from the analysis.
Statistical Analysis
Chi-square and Student t-test were conducted to compare selected characteristics between gastric cancer cases and controls. Difference in selected characteristics that were sex, age, H. pylori infection, CagA and VacAseropositivity, cigarette smoking, alcohol drinking, and gastritis history between cases and controls were determined by a p-value of 0.05.
Hardy-Weinberg equilibrium (HWE) was evaluated in the control group for all SNPs using the chi-square test or Fisher’s exact test with a cut-off level of HWE p-value<0.0001. In the discovery analysis, the association between individual SNPs and gastric cancer risk was evaluated based on raw and permutated p-values using the likelihood ratio test (LRT) with one degree of freedom in the additive, dominant, and recessive models. The additive model assumes a dose response effect with an increasing number of variant alleles. The dominant and recessive models are tests for the minor allele. If d is the minor allele and D is the major allele, the dominant model is DD vs. dd + Dd and the recessive model is dd vs. DD + Dd. Permutated p-values were estimated by 100,000 permutation tests in the single SNP model. To avoid spurious associations with false positive outcomes, the false discovery rate (FDR) using a Benjamini-Hochberg Method was computed [30]. Gastric cancer risk was calculated as odds ratios (ORs) and 95% confidence intervals (CIs) using unconditional logistic regression model adjusting for risk factors that were age, smoking status (ever vs. never), H. pylori infection (positive vs. negative) and CagA seropositivity (positive vs. negative) in the additive, dominant, and recessive models. Haploblocks were created using the default algorithm [31] and tag-SNPs were identified in Haplotype analysis.
In the extension phase, the most significant SNPs in the discovery phase were re-analyzed. Based on the additive and/or recessive models, gastric cancer risk was estimated as OR [95% CI] using unconditional logistic regression model adjusting for the same risk factors as mentioned above. The statistical significance level for the discovery and extension phases was p-value<0.05. To summarize the results from the discovery and extension analyses, pooled- and meta-analyses were conducted. Using the fixed effect model, summarized OR [95% CI] were computed. Also, heterogeneity across the studies was evaluated by the Cochran Q statistics [32].
All statistical analyses were performed using SAS software version 9.1 (SAS Institute, Cary, North Carolina), PLINK software version 1.06 (http://pngu.mgh.harvard.edu/purcell/plink) [33], and Haploview 4.1 software (http:www.broadinstitute.org/haploview/haploview).
Results
There was no significant difference between cases and controls for all selected characteristics in the discovery and extension subjects (p>0.05) (Table 1). In the pooled-analysis, a greater number of cases were CagA and VacA seropositive and smokers (p<0.05).
Table 1. Selected characteristics of gastric cancer cases and controls in the genetic analysis.
| Discovery phase | Extension phase | Total | |||||
| Case(n = 76)N (%) | Control(n = 322)N (%) | Case(n = 386)N (%) | Control(n = 348)N (%) | Case(n = 462)N (%) | Control(n = 670)N (%) | p-value | |
| Age a | 64.5 (8.6) | 62.8 (8.4) | 61.5 (10.5) | 63.1 (8.4) | 62.0 (10.3) | 63.0 (8.4) | 0.10 |
| Female | 20 (26.3) | 98 (30.4) | 130 (33.7) | 110 (31.6) | 150 (32.5) | 208 (31.0) | 0.61 |
| H.pylori infection (+) | 64 (84.2) | 271 (84.2) | 342 (88.6) | 299 (85.9) | 406 (87.9) | 570 (85.0) | 0.18 |
| CagA (+) | 65 (85.5) | 273 (84.8) | 355 (92.0) | 308 (88.5) | 420 (90.9) | 581 (86.7) | 0.03 |
| VacA (+) | 44 (57.9) | 171 (53.1) | 271 (70.2) | 233 (67.0) | 315 (68.2) | 404 (60.6) | <0.01 |
| Ever smoker b | 52 (68.4) | 183 (56.8) | 238 (61.7) | 191 (54.9) | 290 (62.8) | 374 (55.8) | 0.02 |
| Ever drinker c | 46 (60.5) | 184 (57.1) | 239 (62.1) | 206 (59.2) | 285 (61.8) | 390 (58.2) | 0.22 |
| Gastric ulcer history (+) | 8 (10.5) | 25 (7.8) | 59 (17.6) | 50 (18.7) | 67 (17.8) | 75 (16.8) | 0.72 |
Mean (SD); median age for total cases and controls was 62.6 years. Age ranged from 29 to 85 years old.
Ever smokers were defined as former and current smokers.
Ever drinkers were defined as former and current drinkers.
P-value>0.05 for all selected characteristics in the discovery and extension phases.
Of the 108 SNPS in five steroid hormone biosynthesis related genes and PGR analyzed in the discovery phase, 23 SNPs in PGR in the recessive model and 10 SNPs in CYP19A1 in the recessive or additive models were significantly associated with increased gastric cancer risk in the single SNP analysis (p<0.05). PGR rs542384, PGR rs543215, PGR rs613120, and PGR rs1456765 presented 100,000 permutation test p<0.01, although FDR p-values were not significant (Table 2).
Table 2. Association of significant SNPs in the steroid hormone biosynthesis pathway and PGR with gastric cancer risk (discovery phase).
| Chra | Gene | db SNP ID | MAF (%)b | p global c | OR (95% CI)e | p permutation f | ||
| Dominant | Recessive | Additive | ||||||
| 11 | PGR d | rs484389g | C (18.22) | 0.0213 | 1.21 (0.71–2.07) | 3.06 (1.02–9.19) | 1.33 (0.86–2.06) | 0.0342 |
| rs500760 | G (18.22) | 0.0213 | 1.21 (0.71–2.07) | 3.06 (1.02–9.19) | 1.34 (0.87–2.07) | 0.0342 | ||
| rs499699 | G (18.26) | 0.0217 | 1.21 (0.71–2.06) | 3.05 (1.02–9.16) | 1.33 (0.86–2.06) | 0.0342 | ||
| rs563656 | C (18.14) | 0.0217 | 1.22 (0.72–2.08) | 3.05 (1.02–9.16) | 1.34 (0.87–2.08) | 0.0342 | ||
| rs523630 | T (18.34) | 0.0358 | 1.21 (0.71–2.07) | 2.74 (0.93–8.05) | 1.32 (0.86–2.04) | 0.0477 | ||
| rs572402 | G (18.22) | 0.0213 | 1.21 (0.71–2.07) | 3.06 (1.02–9.19) | 1.34 (0.87–2.07) | 0.0342 | ||
| rs511484 | G (18.22) | 0.0213 | 1.21 (0.71–2.07) | 3.06 (1.02–9.19) | 1.34 (0.87–2.07) | 0.0342 | ||
| rs526487 | T (18.22) | 0.0213 | 1.21 (0.71–2.07) | 3.06 (1.02–9.19) | 1.34 (0.87–2.07) | 0.0342 | ||
| rs547378 | A (18.26) | 0.0363 | 1.22 (0.72–2.08) | 2.74 (0.94–8.02) | 1.33 (0.86–2.04) | 0.0480 | ||
| rs11224575 | G (18.39) | 0.0197 | 1.22 (0.71–2.08) | 3.12 (1.04–9.36) | 1.35 (0.87–2.08) | 0.0129 | ||
| rs518382 | T (18.31) | 0.037 | 1.22 (0.71–2.07) | 2.72 (0.93–7.97) | 1.32 (0.86–2.04) | 0.0477 | ||
| rs508533 | A (16.08) | 0.0118 | 1.29 (0.75–2.24) | 3.99 (1.10–14.46) | 1.42 (0.90–2.26) | 0.0216 | ||
| rs542384 | A (15.99) | 0.0109 | 1.23 (0.71–2.14) | 4.00 (1.10–14.47) | 1.38 (0.86–2.19) | 0.0038 | ||
| rs491893 | A (16.21) | 0.0118 | 1.27 (0.73–2.21) | 3.99 (1.10–14.46) | 1.41 (0.89–2.24) | 0.0216 | ||
| rs543215 | A (15.91) | 0.0111 | 1.24 (0.71–2.16) | 3.98 (1.10–14.42) | 1.38 (0.87–2.21) | 0.0074 | ||
| rs613120 | C (15.91) | 0.0111 | 1.24 (0.71–2.16) | 3.98 (1.10–14.42) | 1.38 (0.87–2.21) | 0.0074 | ||
| rs1456764g | A (15.70) | 0.0118 | 1.36 (0.78–2.35) | 3.92 (1.08–14.26) | 1.47 (0.93–2.33) | 0.0216 | ||
| rs1456765 | T (15.74) | 0.0109 | 1.38 (0.80–2.40) | 4.08 (1.13–14.77) | 1.50 (0.94–2.38) | 0.0032 | ||
| rs7106686 | A (15.83) | 0.0118 | 1.33 (0.77–2.31) | 3.99 (1.10–14.46) | 1.46 (0.92–2.31) | 0.0216 | ||
| rs566351 | T (16.25) | 0.0245 | 1.38 (0.80–2.38) | 3.31 (0.96–11.38) | 1.46 (0.93–2.31) | 0.0389 | ||
| rs537681 | T (16.25) | 0.0245 | 1.39 (0.80–2.39) | 3.31 (0.96–11.36) | 1.47 (0.93–2.31) | 0.0389 | ||
| rs501732 | T (18.09) | 0.0241 | 1.17 (0.69–2.01) | 3.32 (0.97–11.41) | 1.30 (0.83–2.05) | 0.0389 | ||
| rs529359 | A (16.16) | 0.0228 | 1.32 (0.76–2.29) | 3.31 (0.96–11.36) | 1.42 (0.90–2.24) | 0.0178 | ||
| 15 | CYP19A1 | rs16964228g | T (13.10) | 0.0511 | 1.68 (0.96–2.93) | 2.95 (0.67–12.94) | 1.64 (1.01–2.66) | 0.0488 |
| rs1902580g | A (16.79) | 0.0328d | 1.05 (0.61–1.81) | 4.84 (1.15–20.33) | 1.25 (0.77–2.02) | 0.0351 | ||
| rs936306g | T (31.06) | 0.0341 | 1.65 (0.97–2.78) | 1.81 (0.79–4.12) | 1.55 (1.04–2.30) | 0.0359 | ||
| rs2470176 | G (31.68) | 0.0295 | 1.62 (0.96–2.74) | 1.86 (0.84–4.12) | 1.52 (1.03–2.24) | 0.0300 | ||
| rs16964254 | G (30.81) | 0.0248 | 1.62 (0.96–2.73) | 2 (0.90–4.43) | 1.54 (1.04–2.27) | 0.0249 | ||
| rs8031463 | C (30.93) | 0.0333 | 1.58 (0.94–2.66) | 1.95 (0.88–4.32) | 1.51 (1.02–2.22) | 0.0348 | ||
| rs10519301 | A (17.05) | 0.0119d | 1.10 (0.64–1.88) | 5.64 (1.47–21.7) | 1.30 (0.81–2.08) | 0.2244 | ||
| rs1004982g | G (29.27) | 0.0372 | 1.56 (0.94–2.61) | 1.91 (0.87–4.22) | 1.51 (1.03–2.21) | 0.0396 | ||
| rs7168331 | C (29.27) | 0.0372 | 1.56 (0.94–2.60) | 1.91 (0.87–4.22) | 1.48 (1.01–2.17) | 0.0401 | ||
| rs1870049 | C (18.34) | 0.0619 | 11.64 (0.98–2.75) | 1.68 (0.51–5.49) | 1.51 (0.99–2.32) | 0.0605 | ||
Chromosome.
Minor allele frequency.
All raw p-values calculated with 1 degree of freedom in additive model except rs1902580, rs10519301.
Rs1902580, rs10519301 raw p-values calculated with 1 degree of freedom in recessive model.
Adjusted for age, smoking, history of H. pylori infection, and CagA infection.
Permutated p-values calculated from 10,000 permutations in the single SNP analysis in the additive model.
SNPs selected for the extension analysis. For PGR, one haploblock created and thus SNP selected according to the following criteria: 1) SNPs on coding or 3UTR region; 2) lower raw p-value and permutated p-value<0.04; 3) tag-SNP using tagger in Haploview. For CYP19A1, six haploblocks created and significant SNPs in discovery phase were located in the blocks 4, 5, and 6. SNP selection for the extension phase was one or more of the following: 1) lower raw p-value and permutated p-value<0.04; 2) tag-SNP using tagger in Haploview.
All FDR p-values>0.05.
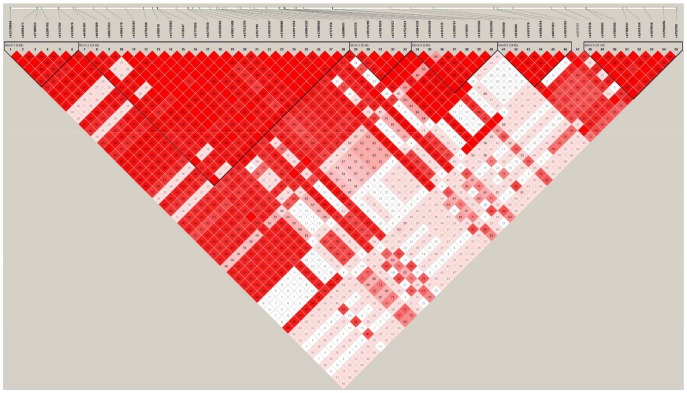
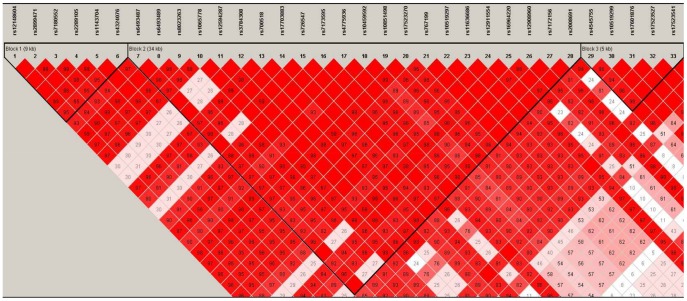
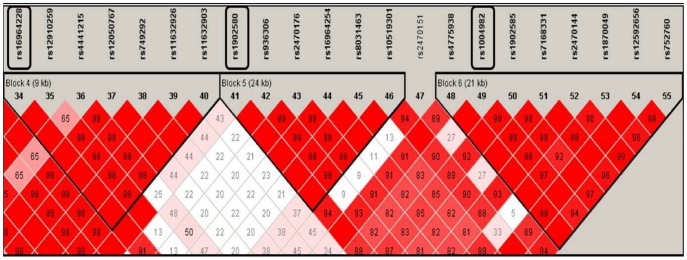
Haplotype blocks were identified by the LD plot. One block was defined by PGR that included all 27 PGR SNPs from the discovery phase (Figure S1), while six blocks were defined by CYP19A1 (Figures 1,2, and 3).
Figure 1. CYP19A1 gene map and LD block.
D’ and LOD values were used for selection of LD color scheme in the discovery phase. Of the six blocks in CYP19A1, significant SNPs in the discovery phase were located in blocks 4, 5, and 6.
Figure 2. CYP19A1 gene map blocks 1, 2, and 3.
Figure 3. CYP19A1 gene map blocks 4, 5, and 6.
Significant SNPs in the discovery phase were located in blocks 4, 5, and 6. SNPs indicated in boxes represent SNPs re-analyzed in the extension phase.
The minor allele frequencies of the SNPs in both the discovery and extension phases were not statistically different. Pooled- and meta-analyses showed CYP19A1 was statistically significantly associated with gastric cancer risk. Minor alleles G, T, and A for rs1004982, rs16964228, and rs1902580, respectively, reported a 1.22 (95% CI [1.01–1.48]), 1.31 (95% CI [1.03–1.66]), and 3.03 (95% CI [1.12–8.18]) increased risk for gastric cancer, respectively, in the pooled-analysis. Meta-analysis showed similar associations. In contrast, all PGR SNPs were not statistically significantly associated with gastric cancer risk (Table 3).
Table 3. Association of most significant SNPs in the steroid hormone biosynthesis pathway with gastric cancer risk in the pooled- and meta-analyses.
| Gene | Chra | SNP | Minorallele | MAFb | Genetic model | OR (95% CI) | |||
| Discovery | Extension | Pooled | Pooled-analysisc | Meta-analysisd | |||||
| CYP19A1 | 15 | rs1004982 | G | 0.2764 | 0.2796 | 0.2780 | Additive | 1.22 (1.01–1.48) | 1.25 (1.02–1.52) |
| Dominant | 1.27 (1.00–1.61) | 1.25 (0.97–1.62) | |||||||
| Recessive | 1.40 (0.91–2.17) | 1.53 (0.97–2.42) | |||||||
| rs16964228 | T | 0.1196 | 0.1371 | 0.1287 | Additive | 1.31 (1.03–1.66) | 1.27 (0.99–1.63) | ||
| Dominant | 1.34 (1.03–1.75) | 1.27 (0.96–1.68) | |||||||
| Recessive | 1.57 (0.72––3.45) | 1.61 (0.72–3.62) | |||||||
| rs1902580 | A | 0.1621 | 0.1653 | 0.1636 | Additive | 1.27 (1.00–1.61) | 1.25 (0.97–1.62) | ||
| Dominant | 0.93 (0.72–1.20) | 0.91 (0.70–1.19) | |||||||
| Recessive | 3.03 (1.12–8.18) | 4.10 (1.43–11.8) | |||||||
Chromosome.
Minor allele frequency.
Adjusted for age, smoking, history of H. pylori infection, and CagA infection.
No heterogeneity (Cochran Q test, P-heterogeneity>0.05) except rs1456764 (p = 0.027) in the recessive mode.
Discussion
CYP19A1 genetic polymorphisms, specifically rs1004982, rs16964228, rs1902580, were associated with an increased risk for gastric cancer in the current study. The discovery analysis showed 23 SNPs in PGR were associated with increased gastric cancer risk and created one large haploblock in haplotype analysis, although associations were not significant in the pooled-analysis.
CYP19A1 encodes CYP19 aromatase, a member of the cytochrome P450 superfamily that is the main enzyme that catalyzes the final and rate-limiting step of estrogen biosynthesis (aromatization of androstenedione and testosterone to estrone and estradiol, respectively) [34]. CYP19 gene is mapped to chromosome 15q21.1, spans about 123 kb, and the regulatory region contains at least 190 distinct promoters that regulate in a signal pathway-specific manner [35] or tissue-specific with hormonally controlled promoters such as gonadal or adipose stroma [36]–[39]. CYP19 mutations have demonstrated increased or decreased aromatase activity thereby altering levels of circulating estrogen [40]–[43]. CYP19A1 genetic variation related studies investigated the association with various hormone related cancers such as breast, endometrial, ovarian, and prostate [44]–[49]. Aromatase activity stimulated breast cancer cell growth [50], aromatase expression levels increased in breast tumors [51], and was the main source of 17β-estradiol in breast tumors and surrounding tissues in postmenopausal women [52], [53]. Studies on the role of CYP19A1 specific to human gastric carcinogenesis are limited. However, strong expression of mRNA CYP19 aromatase was shown in gastric mucosa in adult rats, and aromatase activity in gastric carcinoma human specimens was demonstrated [25]. This suggests a mechanism that polym encoding orphic variants of CYP19 genes may affect cancer susceptibility by altering its encoded enzyme, either through expression or function, to modulate estrogen synthesis. Our findings suggest the possibility that genetic variants of CYP19A1 (rs1004982, rs16964228, and rs1902580) might be involved in altering estrogen levels and affecting apoptosis, mucosal function, carcinogenesis, and thus gastric cancer risk.
There are limited studies that examine genes of the steroid hormone metabolism pathway and gastric cancer. A Japanese study observed a statistically significant association between several CYP19A1 SNPs (rs4646 and rs1902586) and gastric cancer risk [54]. A population-based study in Poland that included 295 gastric cancer cases and 415 controls also genotyped a couple of the same SNPs (rs4646 and rs1902586), however, a significant association was not found [55]. These SNPs were not genotyped in our study, however, other SNPs of CYP19A1 showed statistically significant associations. We genotyped CYP19A1 rs16964228, rs1902580, and rs1004982 that are located in the intron region in three blocks, Block 4, Block 5, and Block 6, respectively. Although the functional relevance of CYP19A1 rs16964228, rs1902580, and rs1004982 for CYP19 enzyme is not clear, CYP19A1 may act as a key marker of individual susceptibility and its genetic variants can modify the development of gastric cancer, but further confirmation is warranted.
Many studies examined CYP19A1 with hormonally associated cancers, such as breast, prostate, endometrial [56]–[59]. SNPs rs10046 (T) and rs936306 (T) are suggested to be ‘high activity alleles’ due to their association with 10% to 20% increased levels of circulating estradiol and estrone in postmenopausal women [58], [60], [61], although did not show significant association with breast cancer [60], [62]. In the current study, rs10046 was not genotyped, but rs936306 was genotyped. While rs936306 was significant in the discovery phase, rs936306 was insignificant in the pooled and meta-analyses.
Our discovery analysis showed 23 SNPs in PGR were associated with an increased risk for gastric cancer. In our haplotype analysis, the significant 23 SNPs from the discovery analysis, in addition to the remaining four PGR SNPs that were genotyped, formed one large block, suggesting these SNPs are correlated with each other and are associated with gastric cancer. However, due to insufficient power in the recessive model, the extension phase did not report a statistically significant association with any PGR SNPs. PGR levels were significantly increased in gastric cancer patients’ tissues while not in normal tissue [63] suggesting gastric mucosa may be the target tissue for progesterone action [64]. Therefore, polymorphic variants of PGR may be involved in modification of gastric cancer susceptibility by altering its encoded receptor status expression and function.
Although this was a two phase study that aimed to increase the number of study subjects, the power was nevertheless low, and did not allow stratified analysis according to hormone related factors such as menopausal status, gender, and cancer type such as cardiac and non-cardiac. The etiology of gastric cancer is multi-factorial, and an in-depth understanding of risk and protective factors and its interactions will help provide an even better understanding of the disease. Moreover, in the extension phase, hospital and community-based cases were matched to community-based controls that may introduce bias. However, information bias was minimized since people are born with their genes and changes in genes are not common. Also, selection bias was minimized because cases were matched to controls according to important risk factors in the initial study design stage.
The study is a two-phase genetic association study. In the candidate approach genetic analysis, significant SNPs that were identified in the discovery phase were re-analyzed in the extension phase. Second, this population-based nested case-control study is free of many biases common in retrospective designs. Confounding factors were adjusted for in multivariate models.
In summary, this population-based two-phase genetic association study reports CYP19A1 genetic variants, rs16964228, rs1902580, and rs1004982, are significantly associated with gastric cancer risk and appear to be a genetic marker of susceptibility in gastric carcinogenesis in the Korean population. Given CYP19A1’s key role in estrogen biosynthesis, CYP19A1 polymorphisms that alter estrogen production can be involved in gastric carcinogenesis. Future studies of estrogen and testosterone biomarkers from blood and urine are needed to confirm and further understand the molecular basis.
Supporting Information
PGR gene map and LD block. D’ and LOD values were used for selection of LD color scheme in the discovery phase. SNPs indicated in boxes represent SNPs re-analyzed in the extension.
(TIFF)
Detailed information on the candidate genes and SNPs in the steroid hormone biosynthesis pathway and PGR .
(DOCX)
Funding Statement
This study was supported by a grant from (1) the National R&D Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea (0520140); (2) the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology [NRF-2009-353-0066258]; and (3) the Basic Research Laboratory (BRL) program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology [2011-0001564]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Parkin DM, Bray F, Ferlay J, Pisani P (2005) Global cancer statistics, 2002. CA Cancer J Clin 55: 74–108. [DOI] [PubMed] [Google Scholar]
- 2. Sipponen P, Correa P (2002) Delayed rise in incidence of gastric cancer in females results in unique sex ratio (M/F) pattern: etiologic hypothesis. Gastric Cancer 5: 213–219. [DOI] [PubMed] [Google Scholar]
- 3.Ferlay J SH, Bray F, Forman D, Mathers C, Parkin DM (2010) GLOBOCAN 2008, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 10. Lyon, France: International Agency for Research on Cancer.
- 4. Freedman ND, Chow WH, Gao YT, Shu XO, Ji BT, et al. (2007) Menstrual and reproductive factors and gastric cancer risk in a large prospective study of women. Gut 56: 1671–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Duell EJ, Travier N, Lujan-Barroso L, Boutron-Ruault MC, Clavel-Chapelon F, et al. (2010) Menstrual and reproductive factors, exogenous hormone use, and gastric cancer risk in a cohort of women from the European Prospective Investigation Into Cancer and Nutrition. Am J Epidemiol 172: 1384–1393. [DOI] [PubMed] [Google Scholar]
- 6. Lindblad M, Garcia Rodriguez LA, Chandanos E, Lagergren J (2006) Hormone replacement therapy and risks of oesophageal and gastric adenocarcinomas. Br J Cancer 94: 136–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Frise S, Kreiger N, Gallinger S, Tomlinson G, Cotterchio M (2006) Menstrual and reproductive risk factors and risk for gastric adenocarcinoma in women: findings from the canadian national enhanced cancer surveillance system. Ann Epidemiol 16: 908–916. [DOI] [PubMed] [Google Scholar]
- 8. Heuch I, Kvale G (2000) Menstrual and reproductive factors and risk of gastric cancer: a Norwegian cohort study. Cancer Causes Control 11: 869–874. [DOI] [PubMed] [Google Scholar]
- 9. Palli D, Cipriani F, Decarli A, Galli M, Saieva C, et al. (1994) Reproductive history and gastric cancer among post-menopausal women. Int J Cancer 56: 812–815. [DOI] [PubMed] [Google Scholar]
- 10. La Vecchia C, D’Avanzo B, Franceschi S, Negri E, Parazzini F, et al. (1994) Menstrual and reproductive factors and gastric-cancer risk in women. Int J Cancer 59: 761–764. [DOI] [PubMed] [Google Scholar]
- 11. Kaneko S, Tamakoshi A, Ohno Y, Mizoue T, Yoshimura T (2003) Menstrual and reproductive factors and the mortality risk of gastric cancer in Japanese menopausal females. Cancer Causes Control 14: 53–59. [DOI] [PubMed] [Google Scholar]
- 12. Persson C, Inoue M, Sasazuki S, Kurahashi N, Iwasaki M, et al. (2008) Female reproductive factors and the risk of gastric cancer in a large-scale population-based cohort study in Japan (JPHC study). Eur J Cancer Prev 17: 345–353. [DOI] [PubMed] [Google Scholar]
- 13. Fernandez E, Gallus S, Bosetti C, Franceschi S, Negri E, et al. (2003) Hormone replacement therapy and cancer risk: a systematic analysis from a network of case-control studies. Int J Cancer 105: 408–412. [DOI] [PubMed] [Google Scholar]
- 14. Heuch I, Kvale G (2003) Does breastfeeding affect the risk of gastric cancer? Int J Cancer 106: 982–983. [DOI] [PubMed] [Google Scholar]
- 15. Inoue M, Ito LS, Tajima K, Yamamura Y, Kodera Y, et al. (2002) Height, weight, menstrual and reproductive factors and risk of gastric cancer among Japanese postmenopausal women: analysis by subsite and histologic subtype. Int J Cancer 97: 833–838. [DOI] [PubMed] [Google Scholar]
- 16. La Vecchia C, Negri E, Franceschi S, Parazzini F (1993) Long-term impact of reproductive factors on cancer risk. Int J Cancer 53: 215–219. [DOI] [PubMed] [Google Scholar]
- 17. Furukawa H, Iwanaga T, Koyama H, Taniguchi H (1982) Effect of sex hormones on carcinogenesis in the stomachs of rats. Cancer Res 42: 5181–5182. [PubMed] [Google Scholar]
- 18. Ohtani M, Garcia A, Rogers AB, Ge Z, Taylor NS, et al. (2007) Protective role of 17 beta -estradiol against the development of Helicobacter pylori-induced gastric cancer in INS-GAS mice. Carcinogenesis 28: 2597–2604. [DOI] [PubMed] [Google Scholar]
- 19. Pricci M, Linsalata M, Russo F, Messa C, Amati L, et al. (2001) Effects of 17beta-estradiol administration on apoptosis and polyamine content in AGS cell line. Anticancer Res 21: 3215–3220. [PubMed] [Google Scholar]
- 20. Katoh M (2003) Trefoil factors and human gastric cancer (review). Int J Mol Med 12: 3–9. [PubMed] [Google Scholar]
- 21. Camargo MC, Goto Y, Zabaleta J, Morgan DR, Correa P, et al. (2012) Sex hormones, hormonal interventions, and gastric cancer risk: a meta-analysis. Cancer Epidemiol Biomarkers Prev 21: 20–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karat D, Brotherick I, Shenton BK, Scott D, Raimes SA, et al. (1999) Expression of oestrogen and progesterone receptors in gastric cancer: a flow cytometric study. Br J Cancer 80: 1271–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kominea A, Konstantinopoulos PA, Kapranos N, Vandoros G, Gkermpesi M, et al. (2004) Androgen receptor (AR) expression is an independent unfavorable prognostic factor in gastric cancer. J Cancer Res Clin Oncol 130: 253–258. [DOI] [PubMed] [Google Scholar]
- 24. Takano N, Iizuka N, Hazama S, Yoshino S, Tangoku A, et al. (2002) Expression of estrogen receptor-alpha and -beta mRNAs in human gastric cancer. Cancer Lett 176: 129–135. [DOI] [PubMed] [Google Scholar]
- 25. Ueyama T, Shirasawa N, Numazawa M, Yamada K, Shelangouski M, et al. (2002) Gastric parietal cells: potent endocrine role in secreting estrogen as a possible regulator of gastro-hepatic axis. Endocrinology 143: 3162–3170. [DOI] [PubMed] [Google Scholar]
- 26. Izawa M, Inoue M, Osaki M, Ito H, Harada T, et al. (2008) Cytochrome P450 aromatase gene (CYP19) expression in gastric cancer. Gastric Cancer 11: 103–110. [DOI] [PubMed] [Google Scholar]
- 27. Jacobs E, Watson SA, Ellis IO, Hardcastle JD, Robertson JF (1997) The effect of onapristone, a progesterone antagonist, on the growth of human gastrointestinal cancer xenografts. Eur J Cancer 33: 1130–1135. [DOI] [PubMed] [Google Scholar]
- 28. Yoo KY, Shin HR, Chang SH, Lee KS, Park SK, et al. (2002) Korean Multi-center Cancer Cohort Study including a Biological Materials Bank (KMCC-I). Asian Pac J Cancer Prev 3: 85–92. [PubMed] [Google Scholar]
- 29. Cho LY, Kim CS, Li L, Yang JJ, Park B, et al. (2009) Validation of self-reported cancer incidence at follow-up in a prospective cohort study. Ann Epidemiol 19: 644–646. [DOI] [PubMed] [Google Scholar]
- 30. Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B (Methodological) 57: 289–300. [Google Scholar]
- 31. Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, et al. (2002) The structure of haplotype blocks in the human genome. Science 296: 2225–2229. [DOI] [PubMed] [Google Scholar]
- 32. Hardy RJ, Thompson SG (1998) Detecting and describing heterogeneity in meta-analysis. Stat Med 17: 841–856. [DOI] [PubMed] [Google Scholar]
- 33. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tsuchiya Y, Nakajima M, Yokoi T (2005) Cytochrome P450-mediated metabolism of estrogens and its regulation in human. Cancer Lett 227: 115–124. [DOI] [PubMed] [Google Scholar]
- 35. Bulun SE, Sebastian S, Takayama K, Suzuki T, Sasano H, et al. (2003) The human CYP19 (aromatase P450) gene: update on physiologic roles and genomic organization of promoters. J Steroid Biochem Mol Biol 86: 219–224. [DOI] [PubMed] [Google Scholar]
- 36. Mahendroo MS, Means GD, Mendelson CR, Simpson ER (1991) Tissue-specific expression of human P-450AROM. The promoter responsible for expression in adipose tissue is different from that utilized in placenta. J Biol Chem 266: 11276–11281. [PubMed] [Google Scholar]
- 37. Harada N, Utsumi T, Takagi Y (1993) Tissue-specific expression of the human aromatase cytochrome P-450 gene by alternative use of multiple exons 1 and promoters, and switching of tissue-specific exons 1 in carcinogenesis. Proc Natl Acad Sci U S A 90: 11312–11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Utsumi T, Harada N, Maruta M, Takagi Y (1996) Presence of alternatively spliced transcripts of aromatase gene in human breast cancer. J Clin Endocrinol Metab 81: 2344–2349. [DOI] [PubMed] [Google Scholar]
- 39. Agarwal VR, Bulun SE, Leitch M, Rohrich R, Simpson ER (1996) Use of alternative promoters to express the aromatase cytochrome P450 (CYP19) gene in breast adipose tissues of cancer-free and breast cancer patients. J Clin Endocrinol Metab 81: 3843–3849. [DOI] [PubMed] [Google Scholar]
- 40. Irahara N, Miyoshi Y, Taguchi T, Tamaki Y, Noguchi S (2006) Quantitative analysis of aromatase, sulfatase and 17beta-HSD(1) mRNA expression in soft tissue metastases of breast cancer. Cancer Lett 243: 23–31. [DOI] [PubMed] [Google Scholar]
- 41. Al Sarakbi W, Mokbel R, Salhab M, Jiang WG, Reed MJ, et al. (2006) The role of STS and OATP-B mRNA expression in predicting the clinical outcome in human breast cancer. Anticancer Res 26: 4985–4990. [PubMed] [Google Scholar]
- 42. Wang H, Li Q, Wang T, Yang G, Wang Y, et al. (2011) A common polymorphism in the human aromatase gene alters the risk for polycystic ovary syndrome and modifies aromatase activity in vitro. Mol Hum Reprod 17: 386–391. [DOI] [PubMed] [Google Scholar]
- 43. Sissung TM, Danesi R, Kirkland CT, Baum CE, Ockers SB, et al. (2011) Estrogen receptor alpha and aromatase polymorphisms affect risk, prognosis, and therapeutic outcome in men with castration-resistant prostate cancer treated with docetaxel-based therapy. J Clin Endocrinol Metab 96: E368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ma X, Qi X, Chen C, Lin H, Xiong H, et al. (2010) Association between CYP19 polymorphisms and breast cancer risk: results from 10,592 cases and 11,720 controls. Breast Cancer Res Treat 122: 495–501. [DOI] [PubMed] [Google Scholar]
- 45. Low YL, Li Y, Humphreys K, Thalamuthu A, Darabi H, et al. (2010) Multi-variant pathway association analysis reveals the importance of genetic determinants of estrogen metabolism in breast and endometrial cancer susceptibility. PLoS Genet 6: e1001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kim JY, Lee CS, Kim HO, Jo YH, Lee J, et al. (2009) The association between genetic polymorphisms in CYP19 and breast cancer risk in Korean women. Oncol Rep 22: 487–492. [PubMed] [Google Scholar]
- 47. Yang HP, Gonzalez Bosquet J, Li Q, Platz EA, Brinton LA, et al. (2010) Common genetic variation in the sex hormone metabolic pathway and endometrial cancer risk: pathway-based evaluation of candidate genes. Carcinogenesis 31: 827–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Goodman MT, Lurie G, Thompson PJ, McDuffie KE, Carney ME (2008) Association of two common single-nucleotide polymorphisms in the CYP19A1 locus and ovarian cancer risk. Endocr Relat Cancer 15: 1055–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sonoda T, Suzuki H, Mori M, Tsukamoto T, Yokomizo A, et al. (2010) Polymorphisms in estrogen related genes may modify the protective effect of isoflavones against prostate cancer risk in Japanese men. Eur J Cancer Prev 19: 131–137. [DOI] [PubMed] [Google Scholar]
- 50. Macaulay VM, Nicholls JE, Gledhill J, Rowlands MG, Dowsett M, et al. (1994) Biological effects of stable overexpression of aromatase in human hormone-dependent breast cancer cells. Br J Cancer 69: 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Irahara N, Miyoshi Y, Taguchi T, Tamaki Y, Noguchi S (2006) Quantitative analysis of aromatase mRNA expression derived from various promoters (I.4, I.3, PII and I.7) and its association with expression of TNF-alpha, IL-6 and COX-2 mRNAs in human breast cancer. Int J Cancer 118: 1915–1921. [DOI] [PubMed] [Google Scholar]
- 52. Brodie A, Lu Q, Nakamura J (1997) Aromatase in the normal breast and breast cancer. J Steroid Biochem Mol Biol 61: 281–286. [PubMed] [Google Scholar]
- 53. Simpson ER, Mahendroo MS, Nichols JE, Bulun SE (1994) Aromatase gene expression in adipose tissue: relationship to breast cancer. Int J Fertil Menopausal Stud 39 Suppl 2: 75–83. [PubMed] [Google Scholar]
- 54. Ikeda S, Sasazuki S, Natsukawa S, Shaura K, Koizumi Y, et al. (2008) Screening of 214 single nucleotide polymorphisms in 44 candidate cancer susceptibility genes: a case-control study on gastric and colorectal cancers in the Japanese population. Am J Gastroenterol 103: 1476–1487. [DOI] [PubMed] [Google Scholar]
- 55. Freedman ND, Ahn J, Hou L, Lissowska J, Zatonski W, et al. (2009) Polymorphisms in estrogen- and androgen-metabolizing genes and the risk of gastric cancer. Carcinogenesis 30: 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ashton KA, Proietto A, Otton G, Symonds I, McEvoy M, et al. (2010) Polymorphisms in genes of the steroid hormone biosynthesis and metabolism pathways and endometrial cancer risk. Cancer Epidemiol 34: 328–337. [DOI] [PubMed] [Google Scholar]
- 57. Justenhoven C, Hamann U, Schubert F, Zapatka M, Pierl CB, et al. (2008) Breast cancer: a candidate gene approach across the estrogen metabolic pathway. Breast Cancer Res Treat 108: 137–149. [DOI] [PubMed] [Google Scholar]
- 58. Cai H, Shu XO, Egan KM, Cai Q, Long JR, et al. (2008) Association of genetic polymorphisms in CYP19A1 and blood levels of sex hormones among postmenopausal Chinese women. Pharmacogenet Genomics 18: 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cunningham JM, Hebbring SJ, McDonnell SK, Cicek MS, Christensen GB, et al. (2007) Evaluation of genetic variations in the androgen and estrogen metabolic pathways as risk factors for sporadic and familial prostate cancer. Cancer Epidemiol Biomarkers Prev 16: 969–978. [DOI] [PubMed] [Google Scholar]
- 60. Haiman CA, Dossus L, Setiawan VW, Stram DO, Dunning AM, et al. (2007) Genetic variation at the CYP19A1 locus predicts circulating estrogen levels but not breast cancer risk in postmenopausal women. Cancer Res 67: 1893–1897. [DOI] [PubMed] [Google Scholar]
- 61. Dunning AM, Dowsett M, Healey CS, Tee L, Luben RN, et al. (2004) Polymorphisms associated with circulating sex hormone levels in postmenopausal women. J Natl Cancer Inst 96: 936–945. [DOI] [PubMed] [Google Scholar]
- 62. Canzian F, Cox DG, Setiawan VW, Stram DO, Ziegler RG, et al. (2010) Comprehensive analysis of common genetic variation in 61 genes related to steroid hormone and insulin-like growth factor-I metabolism and breast cancer risk in the NCI breast and prostate cancer cohort consortium. Hum Mol Genet 19: 3873–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wu CW, Chang HM, Kao HL, Lui WY, P’Eng FK, et al. (1992) The nontransformed progesterone and estrogen receptors in gastric cancer. Gastroenterology 102: 1639–1646. [DOI] [PubMed] [Google Scholar]
- 64. Wu CW, Chi CW, Chang TJ, Lui WY, P’Eng FK (1990) Sex hormone receptors in gastric cancer. Cancer 65: 1396–1400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PGR gene map and LD block. D’ and LOD values were used for selection of LD color scheme in the discovery phase. SNPs indicated in boxes represent SNPs re-analyzed in the extension.
(TIFF)
Detailed information on the candidate genes and SNPs in the steroid hormone biosynthesis pathway and PGR .
(DOCX)