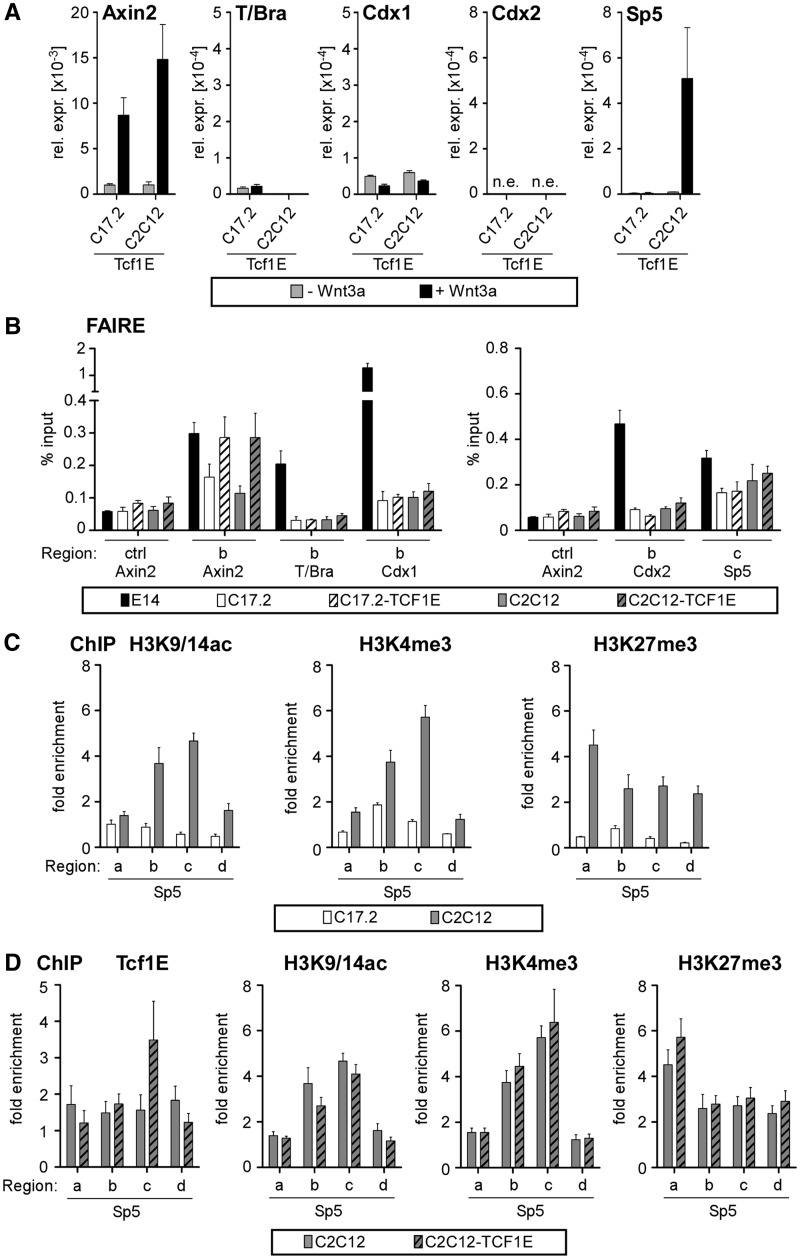
Figure 6.
A pre-existing open chromatin conformation is required for Tcf1E to access its target genes. (A) qRT-PCR analysis of target gene expression after Wnt stimulation in C17.2-Tcf1E and C2C12-Tcf1E cells. The values from all qRT-PCR analyses are the expression levels of each gene normalized to Gapdh. Note the much lower levels of T/Bra and Cdx1 expression compared with E14 ESCs (Figure 1). (B) qPCR after FAIRE analyses to monitor the chromatin conformation states at Axin2, T/Bra, Cdx1, Cdx2 and Sp5 in E14 ESCs, parental C17.2 and C2C12 cells, as well as C17.2-Tcf1E and C2C12-Tcf1E cells. The data are shown as percent input. (C) qChIP with antibodies specific for H3K9/14ac, H3K4me3 and H3K27me3 to determine association with the indicated genomic regions of Sp5 in C17.2 and C2C12 cells. The data are shown as the fold enrichment over Gapdh. (D) qChIP with antibodies specific for HA-tagged Tcf1E, H3K9/14ac, H3K4me3 and H3K27me3 to determine association with the indicated genomic regions for Sp5 in C2C12 and C2C12-Tcf1E cells. The data are shown as the fold enrichment over Gapdh.