Figure 1.
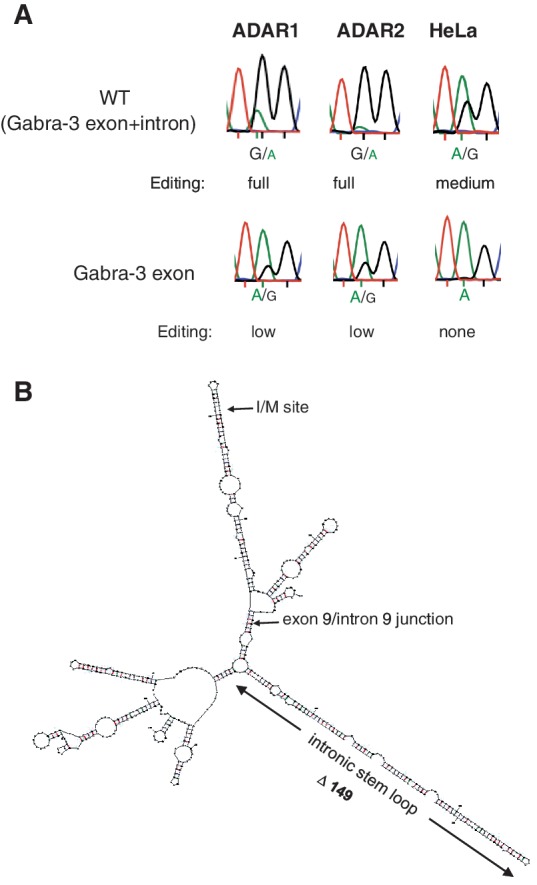
Efficiency of editing at the I/M site using different Gabra-3 constructs. (A) Sanger sequencing results after RT-PCR from co-transfections of ADAR1 or ADAR2 with Gabra-3 editing reporter constructs with or without intronic sequence. Editing is detected as a dual A and G peak at the I/M site and the amount of edited transcripts is determined by measuring the ratio between the A and G peak heights. Reproducible triplicates of these measurements were compared with known levels of editing at the I/M site using 454 HTP sequencing (see ‘Materials and Methods’ section; Supplementary Figure S1). For endogenous editing the reporters were transfected into HeLa cells. (B) Putative RNA secondary structure of the Gabra-3 transcript at exon 9 and part of the intron 9. The edited I/M site as well as the exon/intron junction are indicated. The conserved intronic duplex structure is located 150 nt downstream of the I/M site.
