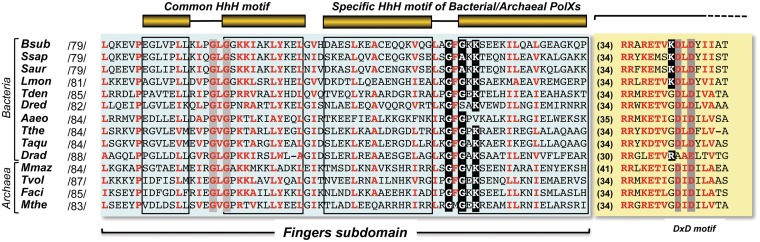
Figure 1.
Multiple amino acid sequences alignment of the fingers subdomain and the DxD motif of the palm subdomain of bacterial/archaeal PolXs. The common HhH motif shared by most of PolX members and the one specifically present in the bacterial/archaeal members are drawn. Numbers between slashes indicate the amino acid position relative to the N-terminus of each DNA polymerase. Numbers in parentheses indicate the length of the intervening amino acid sequence. Because of the large number of sequences, only selected representatives from the Eubacteria and Archea genus are aligned. Names of organisms are abbreviated as follows: Bsub, Bacillus subtilis (GenBank accession number NP_390737); Lmon, Lysteria monocytogenes (GenBank accession number YP_013839); Ssap, Staphylococcus saprolyticus (GenBank accession number YP_301742); Saur, Staphilococcus aureus (GenBank accession number YP_001246578); Dred, Desulfotomaculum reducens (GenBank accession number YP_001112987); Aaeo, Aquifex aeolicus (GenBank accession number NP_213981); Tthe, Thermus thermophilus (GenBank accession number YP_144416); Tden, Thiobacillus denitrificans (GenBank accession number AAZ97399); Mmaz, Methanosarcina mazei (GenBank accession number NP_633918); Faci, Ferroplasma acidarmanus (GenBank accession number ZP_01709777); Mthe, Methanothermobacter thermautotrophicus (GenBank accession number NP_275693); Dred, Deinococcus radiodurans (GenBank accession number NP_294190); Tvol, Thermoplasma volcanium (GenBank accession number NP_111375) and Taqu, Thermus aquaticus (GenBank accession number BAA13425). Conserved residues (≥80% of the aligned polymerases) are indicated with red letters. Conserved glysine and lysine residues of the specific HhH motif and the lysine residue preceding the first catalytic aspartic acid residue are indicated in white letters over a black background. Alignment was made by using the Multalin tool (http://multalin.toulouse.inra.fr/multalin/multalin.html) and was further adjusted by hand.