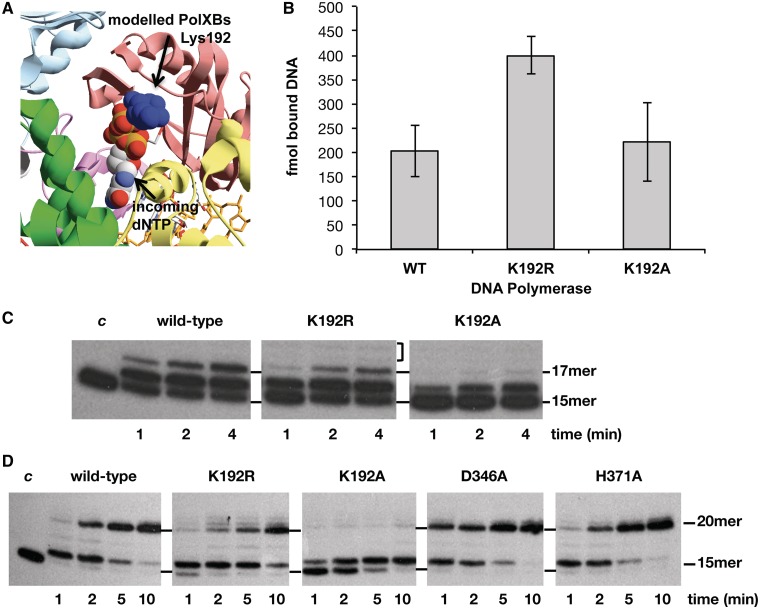
Figure 5.
(A) Potential interaction between PolXBs residue Lys192 and the incoming nucleotide. PolXBs was modelled using as template, the solved structure of PolXDr apoenzyme (PDB 2W9M) (35). Palm subdomains of modelled PolXBs and Polβ ternary complex (PDB 1BPY) (39) were further structurally aligned. (B) Binding of mutant polymerases to a gapped DNA. The experiment was carried out as described in ‘Materials and Methods’ section. Five nanomolar of the 2-nt gapped DNA was incubated with 1 µM of either the wild-type or the indicated mutant PolXBs for 10 minutes at 4°C. The bars chart is the result of three independent experiments. (C) Gap-filling reaction of mutants at residue Lys192 on a 2-nt gapped DNA. The assay was carried out as described in ‘Materials and Methods’ section, incubating 1.5 nM of a 2-nt gapped DNA with 62 nM of either the wild-type or the indicated mutant PolXBs in the presence of 8 mM of MgCl2 and 1 µM of dNTPs for the indicated period at 30°C. The unit length of the primer molecule is indicated. (D) Gap-filling reaction of mutants at residue Lys192 on a 5-nt gapped DNA. The assay was performed essentially as described in (C), using as substrate 1.5 nM of a 5-nt gapped DNA (‘Materials and Methods’).