Abstract
The MspJI modification-dependent restriction endonuclease recognizes 5-methylcytosine or 5-hydroxymethylcytosine in the context of CNN(G/A) and cleaves both strands at fixed distances (N12/N16) away from the modified cytosine at the 3′-side. We determined the crystal structure of MspJI of Mycobacterium sp. JLS at 2.05-Å resolution. Each protein monomer harbors two domains: an N-terminal DNA-binding domain and a C-terminal endonuclease. The N-terminal domain is structurally similar to that of the eukaryotic SET and RING-associated domain, which is known to bind to a hemi-methylated CpG dinucleotide. Four protein monomers are found in the crystallographic asymmetric unit. Analytical gel-filtration and ultracentrifugation measurements confirm that the protein exists as a tetramer in solution. Two monomers form a back-to-back dimer mediated by their C-terminal endonuclease domains. Two back-to-back dimers interact to generate a tetramer with two double-stranded DNA cleavage modules. Each cleavage module contains two active sites facing each other, enabling double-strand DNA cuts. Biochemical, mutagenesis and structural characterization suggest three different monomers of the tetramer may be involved respectively in binding the modified cytosine, making the first proximal N12 cleavage in the same strand and then the second distal N16 cleavage in the opposite strand. Both cleavage events require binding of at least a second recognition site either in cis or in trans.
INTRODUCTION
The control of gene expression in mammals relies in part on the modification status of DNA cytosine residues. DNA cytosine modification is a dynamic process and occurs by converting cytosine (C) to 5-methylcytosine (5mC), established by specific DNA methyltransferases (1,2) and then to 5-hydroxymethylcytosine (5hmC) by Tet (ten–eleven translocation) proteins (3–5). 5mC and 5hmC occur in almost all human tissues and cell types examined (6), but 5hmC is relatively enriched in embryonic stem cells (7) and Purkinje neurons (8). However, our current knowledge of DNA cytosine modification patterns (epigenome) within the defined sequences of the human genome is limited (9). In addition, there are differences between the epigenomes of normal cells and those found during pathologic processes, such as aging, mental health and cancer, among many others (10,11). These still await full documentation.
The MspJI family of modification-dependent restriction endonucleases recognizes hemi-modified 5mC or 5hmC in the context of specific sequences and introduces double-stranded (ds) breaks at fixed distances (N12/N16 from the modified C) (12,13). The sequencing of the digested genomic DNA fragments generated from these endonucleases provides a new method to map modified cytosines in the epigenome (13). However, because there is some sequence specificity in the flanking nucleotides of the modified cytosine, the coverage of the entire epigenome is limited.
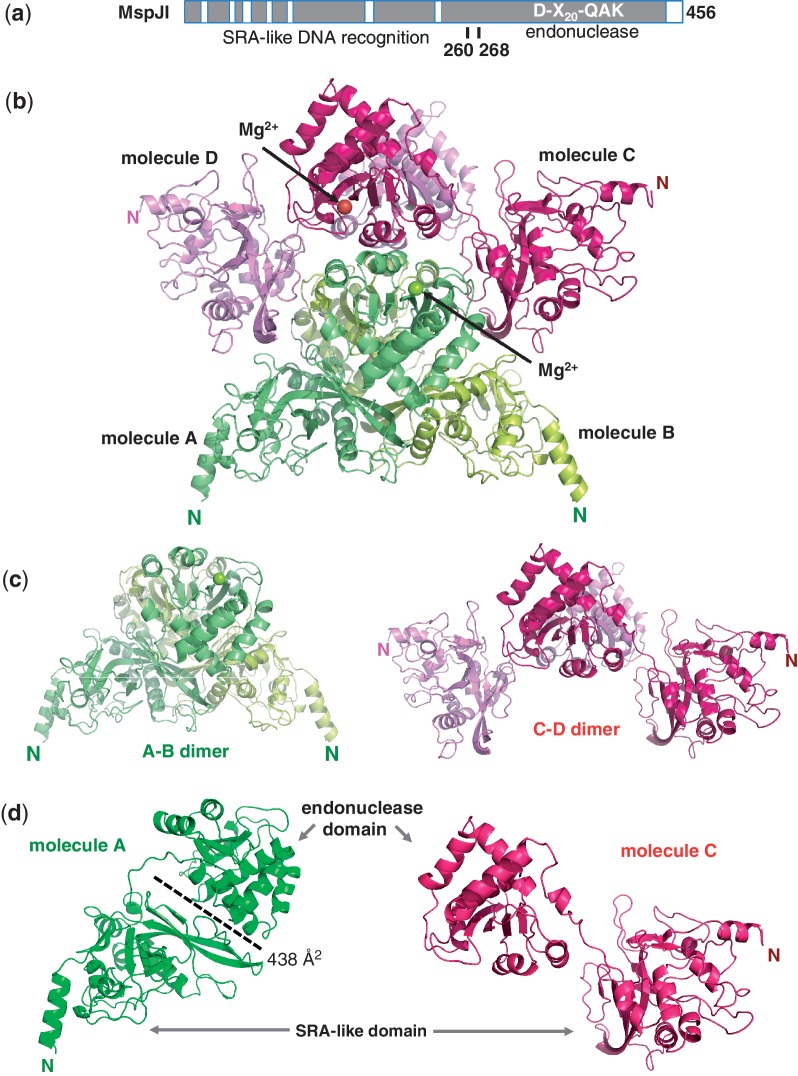
The MspJI family contains at least six characterized members (13). The length of these proteins varies from 388 amino acids in AspBHI to 456 in MspJI, but they all contain a conserved core region of approximately 390 amino acids. MspJI has seven insertions with five to eight residues in the amino-terminal half, mostly in the loop regions and one 15-residue extension in the carboxy-terminus (Figure 1a and Supplementary Figure S1a).
Figure 1.
Structure of MspJI. (a) Schematic representation of MspJI with two domains connected by a linker. A conserved core region is shown in dark grey and the insertions are shown as open boxes (Supplementary Figure S1). (b) Four MspJI monomers, A, B, C and D, form a tetramer. Label ‘N’ indicates amino terminus of each molecule. (c) Two kinds of dimers in closed (A–B dimer) or open (C–D dimer) conformations. (d) Two kinds of monomers with Molecules A or B adopting a closed conformation (in green) and Molecules C or D adopting an open conformation (in red).
As a step to elucidate the mechanism of this unique family of modification-dependent restriction endonucleases, we characterized the structure of MspJI of Mycobacterium sp. JLS. We found that each protein monomer harbors two domains: an amino-terminal SRA-like 5mC DNA-binding domain and a carboxy-terminal endonuclease domain containing the active-site motif of DX20QAK, a variation of the classic PDXn(D/E)XK motif (14–16). Two monomers of MspJI associate to build a primary dimer with two active sites located on opposite faces. These two back-to-back dimers are positioned to form a tetramer with two dsDNA cleavage modules facing opposite directions. Each ds cleavage module contains two active sites facing each other, similar to that of the bona fide dimeric Type II restriction enzymes, enabling dsDNA cuts.
MATERIALS AND METHODS
Protein expression and purification
MspJI wild-type (wt) and mutants were all expressed in T7 Express, i.e. ER2566 (NEB) and purified as described (13). The primers for mutagenesis are listed in Supplementary Table S1. Using the wt pNEB206A-His6MspJI as the template, we did inverse PCR with primer sets containing target mutation sequences. Each 50 µl inverse PCR reaction contained 2 U of the VentR DNA Polymerase (NEB #M0254), 1× ThermoPol Reaction Buffer, 200 μM dNTP Solution Mix (NEB #N0446), 0.9 µM forward and reverse primer, 1% DMSO and 6 ng template DNA. PCR products were purified by spin columns (QIAGEN #28104). Before transformation, the purified PCR products were treated with DpnI for 15 min at 37°C to digest the parental wt sequence. The transformed cells were plated on LB-agar plates containing 100 μg μl−1 ampicillin and incubated at 37°C overnight followed by mini-prep plasmid purification (QIAGEN, Cat. No. 27106). Mutant clones were confirmed by sequencing with M13/pUC sequencing primers (NEB internal S1224 and S1233).
Crystallography
For crystallization, MspJI proteins underwent further purification via tandem HiTrap Q/Heparin (GE Healthcare) and a sizing column HiLoad 16/600 Superdex 200 (GE Healthcare). Final solutions contained 6–20 mg ml−1 protein, 20 mM Tris–HC1 (pH 8.0), 150 mM NaCl, 10% glycerol (v/v), 1 mM ethylenediaminetetraacetic acid (EDTA) and 1 mM dithiothreitol (DTT). Crystallization was carried out by the hanging-drop vapor-diffusion method at 16°C using equal amounts of protein and well solutions.
MspJI crystals were grown under 3–12% polyethylene glycol 3350, 100 mM MgCl2 and 100 mM imidazole (pH 6.2–7.4). Several morphologies of MspJI crystals were observed and seemingly single crystals that diffracted were hemihedrically twinned as reported by the Xtriage component of the program suite PHENIX (17). By far, most crystals were large box-like crystals (which were very birefringent and diffractible), next a population of orthogonal, elongated crystals (not birefringent and did not diffract) and, sometimes, amongst these were a small population of elongated, trigonal crystals (which were birefringent and did diffract).
For phasing studies, a 15 mg ml−1 protein solution of MspJI was exposed to ∼1 mM K2HgI4 at 4°C overnight before crystallization experiments were conducted. This exposure seemed not to hinder crystal production and may have increased growth of the untwinned crystals with the trigonal morphology as untwinned data were collected. The initial map of MspJI was traced utilizing untwinned, Hg-based single-wavelength anomalous data, with Hg positions near at cysteine residues to aid in tracing.
All the data sets were processed using the program HKL2000 (18). Phasing, map production, and model refinement was conducted using the PHENIX software suite (17). Maps and models were visualized with COOT (19) as well as conducting manual model manipulation during refinement rounds.
Analytical ultracentrifugation
Sedimentation velocity analysis was conducted with MspJI at three different concentrations at 20°C and 50 000 rpm using absorbance optics with a Beckman-Coulter XL-I analytical ultracentrifuge. Double sector cells equipped with quartz windows were used. The rotor was equilibrated under vacuum at 20°C and after a period of ∼1 h at 20°C the rotor was accelerated to 50 000 rpm. Absorbance scans at 280 nm were acquired at 4.5-min intervals for ∼6 h. The complete data set was then analyzed using Sedanal (version 5.03) with the model of a monomer/tetramer self-association, plus a non-interacting higher aggregate. These analyses indicated that the MspJI sample, under the experimental conditions, exists as an interacting monomer/tetramer system which is primarily tetrameric (KD of ∼20 nM). There was a small amount (<2%) of higher aggregates present.
MspJI activity titration on the stem-loop oligonucleotide
MspJI activities on the stem-loop oligonucleotide were carried out using a series of 2- or 4-fold titrations of MspJI. The full sequence of the stem-loop oligonucleotide for the top-strand nicking experiment was 5′-GCC ATG CTG TCM AGG CAG GTA GAT GAC GAC CTT (FAM) TTT GGT CGT CAT CTA CCT GCC TGG ACA GCA TGG C-3′ (Integrated DNA Technologies), where M = 5mC. The oligonucleotide was dissolved in water to 10 µM. Each reaction mixture of the titration series consisted of 3 µl NEB buffer 4, 1 µl substrate and a varying amount of MspJI in a total of 30 µl. In the initial reaction, 8.3 µg of MspJI was added, equivalent to an approximate monomeric enzyme to DNA ratio of 16 (or tetramer to DNA ratio of 4). The reaction mixtures were incubated at 37°C for 1 h and resolved on a 20% polyacrylamide gel with 7 M urea. The gel was scanned in the GE Typhoon Variable Mode Imager 9400.
MspJI activity titration on plasmid pBR322 (dcm+)
A series of 4-fold titrations of MspJI were incubated with 200 ng of pBR322 at 37°C for 1 h. The reaction mixtures were treated with proteinase K and run on a 1.2% agarose gel. In the initial reaction, 1.3 µg of MspJI was added, equivalent to a ratio of 64 for monomeric enzyme to C5mCWGG sites (or ratio of 16 for tetramer to 5mC sites).
DNA-binding assay
Varying amounts of MspJI and 0.5 μM of the stem-loop oligonucleotide with an internal fluorescent label were mixed in a 20 μl reaction [20 mM Tris pH 8.2, 60 mM KCl, 5 mM CaCl2, 100 μg/ml BSA, 4% glycerol (v/v) and 1 mM DTT] and incubated for 1 h at 37°C. A 5 μl loading dye (TE in 50% glycerol (v/v) and 0.02% Xylene cyanol) was added per reaction. An 8 μl of the mixture was loaded onto a 6% tris-borate-EDTA (TBE) gel and run at a constant of 150 V in the cold room and then scanned in a GE Typhoon 9400. The TBE gel (Life Techonologies, Cat. No. EC63652) was prepared prior to loading the samples by running in 0.5× TBE buffer at 160 V for 30 min in the cold room.
RESULTS
We crystallized MspJI in two different space groups (P21 and P31), determined the structure by the Hg-based single-wavelength anomalous diffraction phasing method and refined the structures to resolutions of 2.05 Å and 2.8 Å, respectively (Supplementary Table S2). For the structures of both space groups, the crystallographic asymmetric unit contains four molecules, termed Molecules A, B, C and D (Figure 1b). Molecules A and B form a dimer, whereas Molecules C and D form a second dimer (Figure 1c). Two dimers interact to form a tetramer. The two structures, and the respective intra- and inter-molecular interactions, are highly similar with a root mean square deviation of <0.8 Å when comparing 450 pairs of Cα atoms of the Molecule A. Thus we will describe the higher resolution structure of MspJI.
Monomeric MspJI contains two domains (Figure 1d), connected by a linker (residues 260–268) (Figure 1a). Monomers A and B adopt a closed conformation, as the two domains interact with an interface of ∼438 Å2, whereas monomers C and D adopt an open conformation with no direct interactions between the two domains (Figure 1d and Supplementary Figure S1b).
The N-terminal SRA-like DNA-binding domain
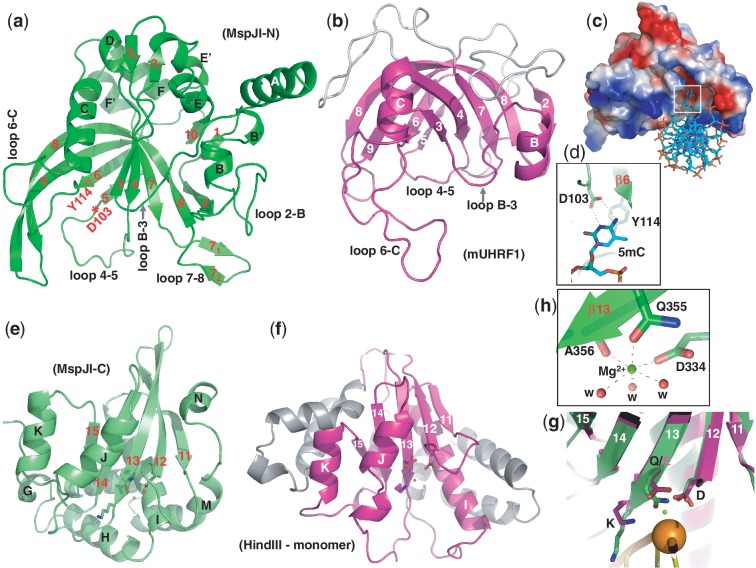
A VAST search (20) against protein structures in the Protein Data Bank (PDB) revealed the N-terminal domain is structurally similar to that of the Arabidopsis SUVH5 SRA domain (21) (P value of 10e − 8.5) and the mammalian UHRF1 SRA domain (P value of 10e − 4.8), which is known to bind to a hemi-methylated CpG dinucleotide (22–24). The SRA-like N-terminal domain contains two twisted β-sheets packed together to form a crescent moon-like structure (Figure 2a). The 20-residue-long curved strand β8 (His175–Asp194) is part of and links together the two β-sheets (Figure 2a). Helix αB is packed against the first β-sheet and helix αC is sandwiched between the two β-sheets. The two helices (αB and αC) and the two sheets, responsible for the crescent-like appearance, are the conserved structural features among the known SRA domains and the N-terminal domain of MspJI (Figure 2b).
Figure 2.
The SRA-like hemi-methylated 5mC recognition domains. (a) Ribbon model of the N-terminal DNA-binding domain of MspJI. In comparison to the SRA domain of mouse UHRF1 (panel b), additional helices of MspJI are positioned on the outer surface of the crescent. In addition, loop 2–B (between strand β2 and helix αB) and loops 7–8 (between strands β7 and β8) in MspJI vary in sequence and length among family members (Supplementary Figure S1a), indicating their potential function in defining the specificity of the recognition of the DNA sequence for the nucleotides flanking of the modified cytosine. (b) The SRA domain of mouse UHRF1 (PDB 3FDE). In mammalian SRA, the corresponding loop between strand β6 and helix αC (6-C) contains residues important for CpG recognition and the loop between helix αB and strand β3 (B-3) for flipping the 5mC out of the DNA helix (22). (c) A surface model of the N-terminal SRA-like DNA recognition domain docked with a DNA containing a flipped 5mC (taken from PDB 3FDE). The surface charge is displayed as blue for positive, red for negative and white for neutral. (d) The flipped 5mC nucleotide can be docked into the binding pocket by interactions (via hydrogen bonds and planar stacking contacts) with Asp103 and Tyr114—two conserved residues among the MspJI family and known SRA domains. Asp103 is part of the loop between strand β4 and β5 (loops 4–5) and the last residue prior to strand β5. Tyr114 is part of the strand β6, which is anti-parallel to strand β5 and sits right next to Asp103. (e) Ribbon model of the C-terminal endonuclease domain of MspJI, which contains five β-strands (β11–β15) and eight helices (αG to αN). Helices αJ, αK, αN are located on one side of the β-sheet and αG, αH, αI, αL, αM on the other side, respectively. (f) The HindIII monomer (taken from the dimer-DNA complex structure; PDB 2E52). (g) Superimposition of the C-terminal endonuclease domain of MspJI (in green) and the HindIII–DNA complex (in magenta) near the scissile phosphate group (shown as an orange ball) (taken from PDB 2E52). Three catalytic residues (Asp334, Gln355 and Lys357 in MspJI and Asp93, Asp108 and Lys110 in HindIII) are shown in a stick model in the carboxyl ends of strands β12 and β13. (h) The octahedral coordination of one Mg2+ observed in the active site.
Five loops, located on the inner surface of the crescent where DNA is bound in the eukaryotic SRA domains, might have functional significance (Figure 2a and b). We created a model of the N-terminal SRA-like domain bound to DNA, by using the coordinates of the mouse SRA–DNA complex (22). After superimposing the protein components, the DNA was positioned over the basic surface of MspJI with an acidic pocket (Figure 2c). In analogy to the SRA–DNA complex, the acidic pocket defines the location of the 5mC-binding site, which is likely to be flipped out from the DNA helix (Figure 2d).
The C-terminal endonuclease domain
The C-terminal domain of MspJI is similar to many structurally characterized endonucleases and other hypothetical proteins (Supplementary Table S3), including HindIII endonuclease (P value of 10e − 6.5), a typical dimeric Type II restriction enzyme (25). The C-terminal endonuclease-like domain contains a central five-stranded β-sheet (β11–β15), flanked by helices (three on one side and five on the other side, respectively) to form an αβα sandwich (Figure 2e). The β-sheet and two pairs of helices on either side of the sheet (αJ and αK, or αI and αL) are structural features conserved between MspJI and HindIII (Figure 2f). The notable missing structural elements in MspJI are the dimerization helices found in HindIII (Supplementary Figure S2a). Using the coordinates of the HindIII–DNA complex, we superimposed the protein components and then positioned the DNA over the C-terminal endonuclease domain of MspJI. The resulting model showed that MspJI could contact the DNA without physical distortion of either the protein or the DNA component. The catalytic residues of HindIII, Asp93, Asp108 and Lys110 align spatially with MspJI residues Asp334, Gln355 and Lys357 of the DX20QAK motif, in which glutamine occurs in place of the second acidic residue (Figure 2g). The side chains of Asp334 and Gln355, the main chain carbonyl oxygen atom of Ala356 and three water molecules coordinate the binding of Mg2+ ion (Figure 2h), which, together with Lys357, cluster around the scissile phosphodiester bond of the docked DNA.
Dimeric form of MspJI
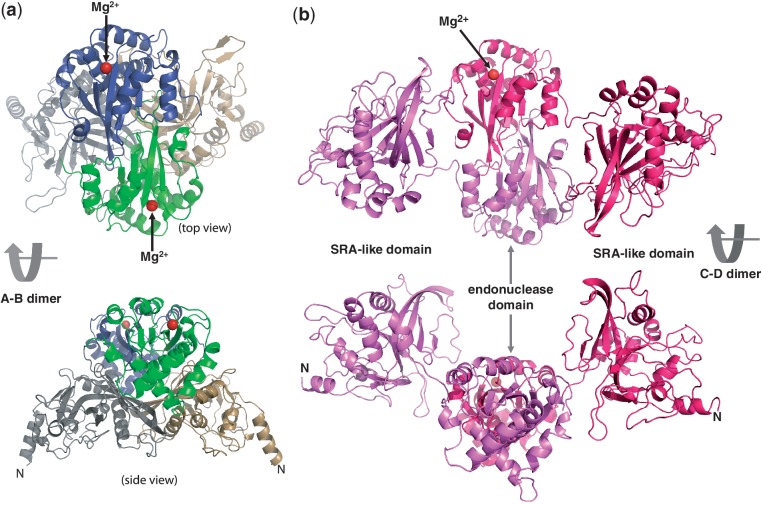
Typical Type II restriction endonucleases, like HindIII, are face-to-face homodimers with two active sites facing each other, and act symmetrically at palindromic DNA sequences, with each active site cutting one strand (Supplementary Figure S2a). We examined all possible protein–protein interfaces of intramolecule interactions among Molecules A to D of MspJI. Among them, the C-terminal endonuclease domains of Molecules A and B, or that of Molecules C and D, form a back-to-back dimer (the interface area of ∼1300 Å2) with two active sites (indicated by the locations of Mg2+) located on opposite faces (Figure 3a and b).
Figure 3.
Two distinct back-to-back dimers of MspJI. (a) Molecules A and B form a closed back-to-back dimer with two active sites (indicated by the Mg2+ sites) located in opposite ends. Molecule A is colored in grey (the N-terminal SRA-like domain) and green (the C-terminal endonuclease domain), while Molecule B is in light orange and dark blue, respectively. (b) Molecules C and D form an open back-to-back dimer with no direct interactions between the two N-terminal DNA-binding domains.
Interestingly, we note that the Molecules A and B (with the closed conformation) are of higher quality with electron densities observed for residues from 5 to 456 continuously, while electron densities for Molecules C and D (with the open conformation) are discontinuous in several loops or missing for many side chains. The difference in crystallographic thermal stability may indicate a relative movement between the two dimers. While the back-to-back dimer interface of Molecules C and D are mainly mediated by the C-terminal endonuclease domain (Figure 3b), both the N- and C-terminal domains of Molecules A and B contribute significantly to the large interface of A–B dimer (∼3800 Å2) (Figure 3a).
To analyze the significance of the dimer interface, we mutated Val191 to charged and/or bulky side chains (V191D and V191R). Val191 of strand β8 sits in the three way junction of the N- and C-domains of Molecule A (or B) as well as the N-terminal domain of Molecule B (or A) (Supplementary Figure S3a). Mutants V191D and V191R have decreased protein yield (Supplementary Figure S3b) and exhibited lower specific activity (Supplementary Figure S3c).
Tetrameric form of MspJI
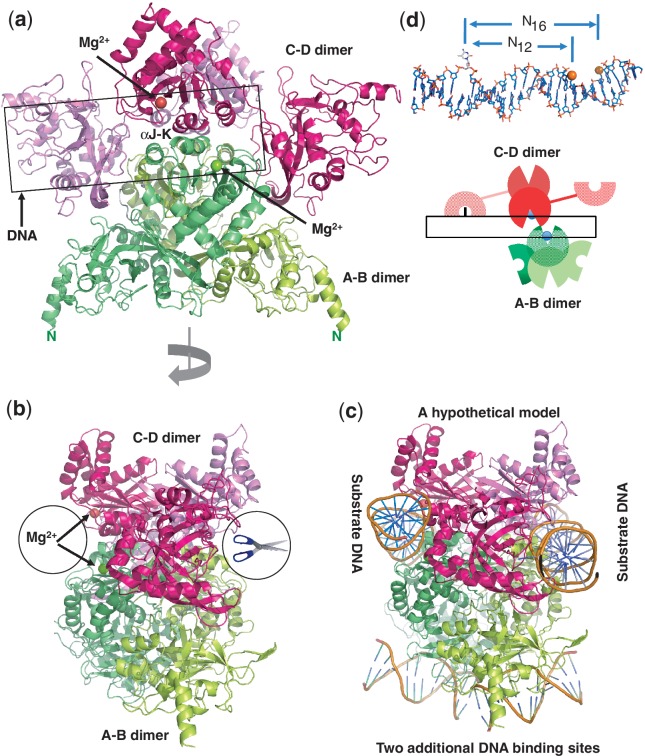
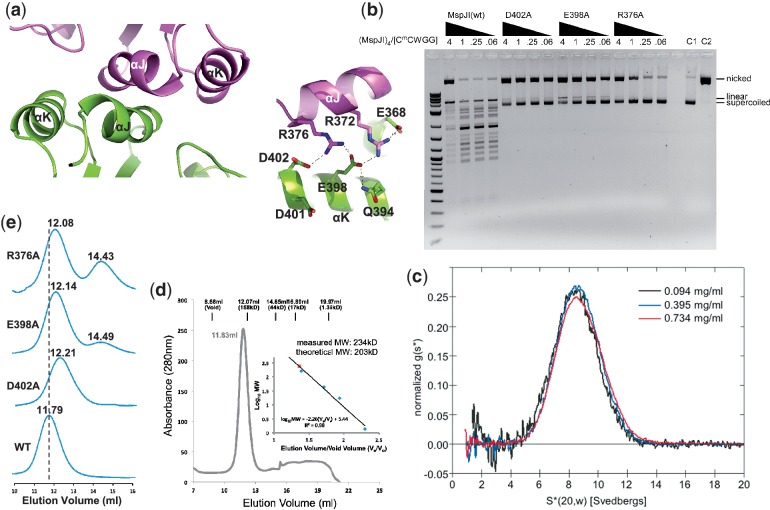
Two back-to-back dimeric units are further dimerized to form a tetramer (Figure 4a), mediated by two pairs of helices αJ and αK of Molecules A and C or Molecules B and D. Such arrangement brings two active sites of Molecules A and C forming a face-to-face architecture (Figure 4b). Helices αJ and αK are rich in charged residues conserved among the family members (Supplementary Figure S1a). Buried in the interface between two pairs of helices αJ and αK is a network of charge–charge intermolecular interactions involving Asp402–Arg376–Glu398–Arg372–Glu368 (Figure 5a) and appears critical for stabilizing the tetramer. Consistent with this hypothesis, substitution of Asp402, Arg376 or Glu398 for alanine (D402A, R376A or E398A) resulted in much reduced ds cleavage activity so that the cleavage stops after the nicking step (Figure 5b). Analytical ultracentrifugation (Figure 5c) and analytical gel-filtration (Figure 5d) measurements confirm that the wild-type protein exists as a tetramer in solution, whereas the mutants displayed a slightly delayed elution peak and a secondary, monomeric peak particularly notable in the case of R376A (Figure 5e).
Figure 4.
A unique tetramer of MspJI. (a) Two back-to-back dimers form a tetramer with four DNA-binding domains and two face-to-face ds ‘scissors’ that cleave hexanucleotides producing four base pair staggers (N12/N16). (b) A 90° view from that of panel a. (c) A hypothetical model of MspJI with two DNA molecules bound in the active sites of the ‘scissors’ (panel b). These two DNA molecules could be connected through DNA looping. Two additional 5mC-containing DNAs could be bound through the N-terminal DNA-binding domains of A–B dimer (bottom). (d) A cartoon illustration of the proposed MspJI tetramer–DNA complex mediated by reading of 5mC by one monomer (Molecule C), cutting the proximal N12 site in the top strand by the second monomer (Molecule D) and the distal N16 cut in the bottom strand by the third monomer (Molecule A). Top panel shows a DNA molecule with a flipped 5mC and the proximal N12 (top strand) and distal N16 (bottom strand) cleavage sites.
Figure 5.
MspJI exists as a tetramer in solution. (a) The major tetramer interface is mediated by helices αJ and αK (left panel), including a network of charge–charge interactions (right panel). (b) Activity profiles of the mutants D402A, E398A and R376A showing the cleavage stalled in the nicked state, compared with the MspJI wild-type. The 4-fold titrations of MspJI and mutants were incubated with 200 ng of pBR322 at 37°C for 2 h. (c) Analytical ultracentrifugation of MspJI at three different concentrations. Scans were taken every 4.5 min and were used to calculate the normalized sedimentation coefficient distribution, g(s*). The tetramer has a sedimentation coefficient corrected to standard conditions, S(20,w), of 8.64 S. (d) Elution profile of MspJI on a Superdex 200 (10/300 GL) (GE Healthcare). The column buffer was 20 mM Tris, pH 8.0, 1 mM EDTA, 10% glycerol (v/v), 1 mM DTT and 150 mM NaCl, and ∼1.7 mg of MspJI was loaded onto the column. The inset shows the standardization of the size exclusion column using a protein marker kit (Biorad) at the time MspJI was profiled using the same buffer. (e) Elution profiles of MspJI mutants (D402A, E398A and R376A) and wt on a Superdex 200 (10/300 GL). The column buffer was the same as in panel d and ∼100 µg of protein was loaded onto the column in four consecutive runs.
Although structures of tetrameric Type IIF restriction enzymes (Cfr10I, Bse634I, NgoMIV and SfiI) have been described previously (26,27) (Supplementary Figure S2b and S2c), there are at least three major differences between the Type IIF tetramers and the MspJI tetramer. First, the polypeptide chains of Type IIF enzymes are folded into a compact single module structure containing both DNA recognition and cleavage functions. In MspJI, two domains connected by a linker appear to independently perform the two functions. Second, two monomers of Type IIF enzymes associate to build a bona fide dimeric restriction enzyme with two active sites located face-to-face. In MspJI, the back-to-back dimer puts the two active sites on the opposite faces, analogous to that of the back-to-back ‘nicking’ endonuclease HinP1I (28) (Supplementary Figure S2d). Third, while different arrangements of two dimers result in two face-to-face (ds) DNA cleavage modules for both tetrameric Type IIF restriction enzymes and the MspJI tetramer (Supplementary Figure S2b and S2e), the MspJI tetramer has the potential to bind two additional DNA molecules (Figure 4c).
MspJI generates a top-strand nicked intermediate
From the primary monomeric structure and the substrate cleavage pattern, MspJI is similar to FokI, which cuts ‘top’ and ‘bottom’ strands 9 and 13 nt (N9/N13) downstream of its non-palindromic recognition sequence. FokI is a monomeric protein with an N-terminal DNA recognition domain that covers the entire recognition sequence and a C-terminal cleavage domain containing one active site (29). To cut both DNA strands, the monomeric FokI bound at the recognition site dimerizes with a second monomer (30), and the initial monomer bound to the recognition site makes the distal N13 cut in the bottom strand, while the recruited monomer makes the proximal N9 cut in the top strand (31). This observation can be explained by the structural requirement of the C-terminal cleavage domain of the initial FokI monomer to relocate to the scissile bond in the bottom strand (32,33). Our initial modeling study of MspJI monomer bound with DNA suggested the same scenario, where the C-terminal endonuclease domain of the same monomer bound to the modified cytosine would make the distal cut in the bottom strand (Supplementary Figure S4).
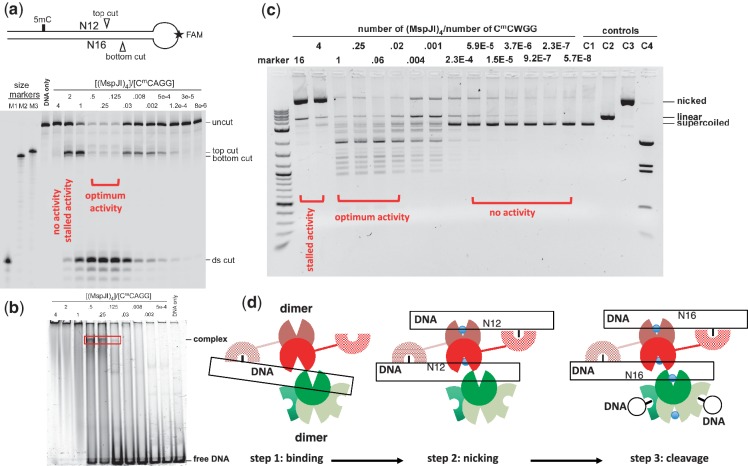
To investigate the cleavage order of the two DNA strands by MspJI, we designed a stem-loop structured oligonucleotide, with a fluorescent label inside the loop (Figure 6a, top panel). The nicked product in the bottom strand would be 4-nt shorter than that in the top strand (Figure 6a, lanes M2 and M3). Titration of increasing amount of MspJI shows the accumulation of nicked top strand (top cut) and ds cleavage (ds cut), but no evidence of nicked bottom strand, suggesting that cleavage happens first in the top strand at the proximal N12 site and then proceeds to the distal N16 site in the bottom strand (Figure 6a). This is inconsistent with the FokI-like model where the same monomer binds 5mC and makes the initial cut on the bottom strand (Supplementary Figure S4), but would be in agreement with the model illustrated in Figure 4d. 5mC DNA binding by one monomer of the back-to-back dimer places the catalytic domain of the other monomer at the top-strand N12 cleavage site, resulting in top-strand nicking. A second cut at the N16 site would require a third monomer from the second back-to-back dimer.
Figure 6.
MspJI cleaves via a top-strand nicked intermediate. (a) A stem-loop structured oligonucleotide substrate containing one 5mC site was designed with an internal fluorescent FAM label in the loop. Size markers were synthesized according to predicted cleavage sites: M1, product from a ds cleavage; M2, product from the bottom-strand cleavage; M3, product from the top-strand cleavage. A 2-fold titration of MspJI digestion started from the tetramer to DNA ratio of 4 to 0.125 and followed by 4-fold dilution. (b) DNA-binding assays were performed by incubating 0.5 µM FAM labeled stem-loop oligonucleotides with varying amount of MspJI tetramer at 37°C for 1 h. (c) MspJI titration on pBR322 (dcm+) containing six C5mCWGG sites. The molar ratio of MspJI tetramer to its substrate sites is shown on the top of the lanes. Control lanes include: C1, pBR322 only; C2, pBR322 digested with EcoRI, which produces linearized pBR322; C3, pBR322 digested with nicking endonuclease Nt.BspQI, which produces a nicked pBR322; C4, pBR322 digested with BstNI, which produces a complete digestion pattern at C5mCWGG. (d) A proposed three-step mechanism of the MspJI enzymatic reaction. Step 1: one SRA-like domain binds specifically to the modified cytosine. With a tetramer-to-DNA ratio of 4:1, no enzymatic activity was observed. Step 2: the other SRA-like domain of the same back-to-back dimer binds another target site, resulting in a top-strand nicked intermediate. With a tetramer-to-DNA ratio of 2:1, the reaction stalled after the first N12 cut. Step 3: with a tetramer-to-DNA ratio of 1:4, the highest level of ds cleavage was observed.
The highest level of cleavage was observed with molar ratios of MspJI tetramer to substrate DNA ranging from 0.5, 0.25 to 0.125 (i.e. monomer-to-DNA ratio of 2, 1 to 0.5) (Figure 6a), suggesting that the most efficient cleavage occurs when all four SRA-like domains of the tetramer are occupied by 5mC DNA. The optimal cleavage activity correlates well with the ability to form a specific complex in electrophoresis mobility-shift assay (Figure 6b, lanes 0.5, 0.25 and 0.125). The amount of this specific complex increases proportionally with the tetramer-to-DNA ratio from 0.125 to 0.5 and disappears when the ratio reaches 1. While the decline in cleavage activity at lower enzyme concentration is expected, surprisingly, further increasing of the MspJI/DNA ratio resulted in a drastic reduction in activity, and no cleavage occurred when the tetramer-to-DNA ratio reaches 4 (i.e. monomer-to-DNA ratio of 16), indicating that more than one DNA molecule must be bound to each tetramer for MspJI cleavage (even the initial nicking) to occur.
We observed a similar phenomenon in the plasmid DNA digestion (Figure 6c). Under a high molar ratio of enzyme to 5mC sites, the digestion of MspJI appears to arrest after the first nicking step on plasmid pBR322 (dcm+) (Figure 6c, lanes with tetramer-to-site ratio of 16 and 4). Further dilution with a tetramer-to-site ratio between 1 and 0.02 rescued such an arrest (Figure 6c), suggesting that the second cleavage event on the other strand requires the tetramer bound to at least another recognition site. Under the high enzyme-to-site ratio (16 and 4), the available free sites may be rare, resulting in impeded activity. It is unclear why nicking can still occur on plasmid DNA, in contrast to oligonucleotide substrates (Figure 6a, lane 5, tetramer-to-DNA ratio of 4). We note that ratio of enzyme to hemi-methylated DNA with the oligonucleotide substrates (Figure 6a) may not be equivalent to that of enzyme to 5mC sites with the fully methylated plasmid substrates (Figure 6c). The supercoiled plasmid DNA and the spatial distance between any two sites in cis may also affect the efficiency of the nicking and cleavage. Nevertheless, the inhibition of a high ratio of enzyme to substrate on activity of tetrameric restriction endonucleases (such as SfiI) had been observed previously (34). It is also known that many type IIF tetrameric enzymes cleave plasmid DNA containing two recognition sites faster than a single site plasmid [reviewed in (26)]. Particularly relevant to our study is that by adding a second DNA with the recognition sequence in trans can accelerate the slow reactions on single-site substrates by the Type IIs tetrameric BspMI, which binds a 6-bp non-palindromic recognition sequence and cleaves the DNA downstream in both strands (35).
DISCUSSION
Here we show, structurally and enzymatically, that MspJI harbors two domains: an SRA-like 5mC-binding domain that recognizes 5mC in the context of CNN(G/A) and an endonuclease domain that cleaves at N12/N16 from the 3′-side of the 5mC. Together with evidence from mammalian and plant SRA domains (36–38), the widespread MspJI-like genes in the bacterial species suggest that they might have evolved different sequence specificities, with some being specific to hemimethylated CpGs while others target 5mC within other sequence contexts. [For a more comprehensive study of domain fusion of a DNA-recognition element to a nuclease, see (39)]. It is interesting to note that DpnI, an N6-methyladenine-dependent Type IIM restriction endonuclease, contains an N-terminal catalytic PDXn(D/E)XK domain and a C-terminal DNA-binding domain (40), in reverse order to the domain arrangement of MspJI. In addition, a recent structural study revealed the isolated N-terminal DNA-binding domain of the 5mC-specific endonuclease McrBC from Escherichia coli flips 5mC as well as an unmodified cytosine in the crystal structure in a sequence independent manner (41).
Unlike monomeric FokI, which shares a similar domain organization and cleaves similarly at an asymmetrical recognition site, MspJI assembles into a tetramer with two dsDNA cleavage modules and two additional DNA-binding domains (Figure 4c). For FokI, the ds cleavage is thought to occur by two interacting monomers bound at the cleavage site (32,33). In comparison, Ecl18kI, exists as a dimer in solution and forms a tetramer while looping a DNA molecule containing two recognition sites (Supplementary Figure 2f) (42,43). Our current working model of the MspJI tetramer reaction involves three sequential steps (Figure 6d): specific binding to the modified cytosine, followed by the first N12 cleavage and then by the second N16 cleavage. Both cleavage events require the binding of at least another recognition site either in cis or in trans.
A commonality between monomeric FokI and tetrameric MspJI is that the DNA-binding events are prerequisites for the cleavage process, which likely activates the MspJI endonuclease domains by controlling the relative movement between the dimers (as seen by different crystallographic thermal factors and the tetramer interface mutants, Figure 5) and resulting in dsDNA cleavage. An MspJI–DNA complex structure will reveal whether any allosteric conformational changes take place upon DNA binding.
ACCESSION NUMBERS
Protein Data Bank: The coordinates and structure factors of MspJI have been deposited with accession numbers 4F0Q (in P21 space group) and 4F0P (in P31 space group).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figures 1–4.
FUNDING
The U.S. National Institutes of Health (NIH) (GM095209 to Y.Z. and GM049245-18 to X.C.) and New England Biolabs. X.C. is a Georgia Research Alliance Eminent Scholar. The Department of Biochemistry at the Emory University School of Medicine supported the use of the Southeast Regional Collaborative Access Team synchrotron beamlines at the Advanced Photon Source of Argonne National Laboratory. Funding for open access charge: NIH.
Conflict of interest statement: The subject of this article is a product of New England Biolabs.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Jeffrey Lary and James Cole at the Biotechnology and Bioservices Center in the University of Connecticut for performing analytical ultracentrifugation analysis of MspJI, Brenda Baker, Nancy Considine and John Buswell at the organic synthesis unit of New England Biolabs for synthesizing the oligonucleotides, Geoffrey Wilson, Hua Wang, Elisabeth Raleigh and William Jack for discussion, Keith Lunen and Daniel Heiter for technical advice. J.R.H. performed crystallographic work, M.Y.M constructed and assessed all the mutants, M.Y.M and D.C.-K. performed nicking and gel shift experiments, D.C.-K. and M.S. performed large-scale purification, X.Z and R.M.G performed purification and early crystallization trials, R.J.R, Y.Z. and X.C. organized and designed the scope of the study, and all were involved in analyzing data and preparing the manuscript. M.Y.M. and D.C.-K. contributed equally.
REFERENCES
- 1.Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol. 1988;203:971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- 2.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 3.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2011;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, Bruckl T, Biel M, Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coppieters N, Dragunow M. Epigenetics in Alzheimer's disease: a focus on DNA modifications. Curr. Pharm. Design. 2011;17:3398–3412. doi: 10.2174/138161211798072544. [DOI] [PubMed] [Google Scholar]
- 11.Irier HA, Jin P. Dynamics of DNA methylation in aging and Alzheimer's Disease. DNA Cell Biol. 2012 doi: 10.1089/dna.2011.1565. February 7 (doi:10.1089/dna.2011.1565; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng Y, Cohen-Karni D, Xu D, Chin HG, Wilson G, Pradhan S, Roberts RJ. A unique family of Mrr-like modification-dependent restriction endonucleases. Nucleic Acids Res. 2010;38:5527–5534. doi: 10.1093/nar/gkq327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen-Karni D, Xu D, Apone L, Fomenkov A, Sun Z, Davis PJ, Kinney SR, Yamada-Mabuchi M, Xu SY, Davis T, et al. The MspJI family of modification-dependent restriction endonucleases for epigenetic studies. Proc. Natl Acad. Sci. USA. 2011;108:11040–11045. doi: 10.1073/pnas.1018448108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pingoud A, Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pieper U, Pingoud A. A mutational analysis of the PD … D/EXK motif suggests that McrC harbors the catalytic center for DNA cleavage by the GTP-dependent restriction enzyme McrBC from Escherichia coli. Biochemistry. 2002;41:5236–5244. doi: 10.1021/bi0156862. [DOI] [PubMed] [Google Scholar]
- 16.Kosinski J, Feder M, Bujnicki JM. The PD-(D/E)XK superfamily revisited: identification of new members among proteins involved in DNA metabolism and functional predictions for domains of (hitherto) unknown function. BMC Bioinformatics. 2005;6:172. doi: 10.1186/1471-2105-6-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otwinowski Z, Borek D, Majewski W, Minor W. Multiparametric scaling of diffraction intensities. Acta Crystallogr. A. 2003;59:228–234. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- 19.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 20.Gibrat JF, Madej T, Bryant SH. Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- 21.Rajakumara E, Law JA, Simanshu DK, Voigt P, Johnson LM, Reinberg D, Patel DJ, Jacobsen SE. A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo. Genes Dev. 2011;25:137–152. doi: 10.1101/gad.1980311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 24.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe N, Takasaki Y, Sato C, Ando S, Tanaka I. Structures of restriction endonuclease HindIII in complex with its cognate DNA and divalent cations. Acta Crystallogr. D Biol. Crystallogr. 2009;65:1326–1333. doi: 10.1107/S0907444909041134. [DOI] [PubMed] [Google Scholar]
- 26.Siksnys V, Grazulis S, Huber R. Structure and function of the tetrameric restriction enzymes. Nucleic Acids Mol. Biol. 2004;14:237–259. [Google Scholar]
- 27.Vanamee ES, Viadiu H, Kucera R, Dorner L, Picone S, Schildkraut I, Aggarwal AK. A view of consecutive binding events from structures of tetrameric endonuclease SfiI bound to DNA. EMBO J. 2005;24:4198–4208. doi: 10.1038/sj.emboj.7600880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horton JR, Zhang X, Maunus R, Yang Z, Wilson GG, Roberts RJ, Cheng X. DNA nicking by HinP1I endonuclease: bending, base flipping and minor groove expansion. Nucleic Acids Res. 2006;34:939–948. doi: 10.1093/nar/gkj484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wah DA, Bitinaite J, Schildkraut I, Aggarwal AK. Structure of FokI has implications for DNA cleavage. Proc. Natl Acad. Sci. USA. 1998;95:10564–10569. doi: 10.1073/pnas.95.18.10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc. Natl Acad. Sci. USA. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanders KL, Catto LE, Bellamy SR, Halford SE. Targeting individual subunits of the FokI restriction endonuclease to specific DNA strands. Nucleic Acids Res. 2009;37:2105–2115. doi: 10.1093/nar/gkp046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wah DA, Hirsch JA, Dorner LF, Schildkraut I, Aggarwal AK. Structure of the multimodular endonuclease FokI bound to DNA. Nature. 1997;388:97–100. doi: 10.1038/40446. [DOI] [PubMed] [Google Scholar]
- 33.Vanamee ES, Santagata S, Aggarwal AK. FokI requires two specific DNA sites for cleavage. J. Mol. Biol. 2001;309:69–78. doi: 10.1006/jmbi.2001.4635. [DOI] [PubMed] [Google Scholar]
- 34.Szczelkun MD, Halford SE. Recombination by resolvase to analyse DNA communications by the SfiI restriction endonuclease. EMBO J. 1996;15:1460–1469. [PMC free article] [PubMed] [Google Scholar]
- 35.Gormley NA, Hillberg AL, Halford SE. The type IIs restriction endonuclease BspMI is a tetramer that acts concertedly at two copies of an asymmetric DNA sequence. J. Biol. Chem. 2002;277:4034–4041. doi: 10.1074/jbc.M108442200. [DOI] [PubMed] [Google Scholar]
- 36.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 37.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 38.Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr. Biol. 2007;17:379–384. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grazulis S, Manakova E, Roessle M, Bochtler M, Tamulaitiene G, Huber R, Siksnys V. Structure of the metal-independent restriction enzyme BfiI reveals fusion of a specific DNA-binding domain with a nonspecific nuclease. Proc. Natl Acad. Sci. USA. 2005;102:15797–15802. doi: 10.1073/pnas.0507949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siwek W, Czapinska H, Bochtler M, Bujnicki J, Skowronek K. Crystal structure and mechanism of action of the N6-methyladenine-dependent type IIM restriction endonuclease R.DpnI. Nucleic Acids Res. 2012;40:7563–7572. doi: 10.1093/nar/gks428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sukackaite R, Grazulis S, Tamulaitis G, Siksnys V. The recognition domain of the methyl-specific endonuclease McrBC flips out 5-methylcytosine. Nucleic Acids Res. 2012;40:6850–6862. doi: 10.1093/nar/gks332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaremba M, Owsicka A, Tamulaitis G, Sasnauskas G, Shlyakhtenko LS, Lushnikov AY, Lyubchenko YL, Laurens N, van den Broek B, Wuite GJ, et al. DNA synapsis through transient tetramerization triggers cleavage by Ecl18kI restriction enzyme. Nucleic Acids Res. 2010;38:7142–7154. doi: 10.1093/nar/gkq560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bochtler M, Szczepanowski RH, Tamulaitis G, Grazulis S, Czapinska H, Manakova E, Siksnys V. Nucleotide flips determine the specificity of the Ecl18kI restriction endonuclease. EMBO J. 2006;25:2219–2229. doi: 10.1038/sj.emboj.7601096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.